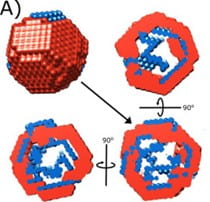
J. San Emeterio, L. Pollack.
Visualizing a viral genome with contrast variation small angle X-ray scattering
J. Biol. Chem. 295(47), 15923-15932 (2020)
G. D. Calvey, A. M. Katz, K. A. Zielinski, B. Dzikovski, L. Pollack
Characterizing Enzyme Reactions in Microcrystals for Effective Mix-and-Inject Experiments using X-ray Free-Electron Lasers
Analytical Chemistry. 92(20), 13864-13870 (2020)
Y. L. Chen, L. Pollack
Machine learning deciphers structural features of RNA duplexes measured with solution X-ray scattering
IUCrJ. 7(5). 870-880 (2020)
J. A. Kelly, A. N. Olson, K. Neupane, S. Munshi, J. San Emeterio, L. Pollack, M. T. Woodside, J. D. Dinman
Structural and functional conservation of the programmed? 1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2)
J. Biol. Chem. 295(31), 10741-10748 (2020)
R. Welty, M. Rau, S. A. Pabit, M. S. Dunstan, G. L. Conn, L. Pollack, K. B. Hall
Ribosomal protein L11 selectively stabilizes a tertiary structure of the GTPase center rRNA domain
J. Mol. Biol. 432(4), 991-1007 (2020)
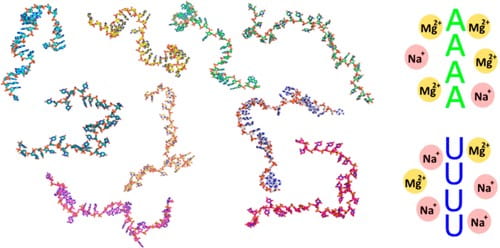
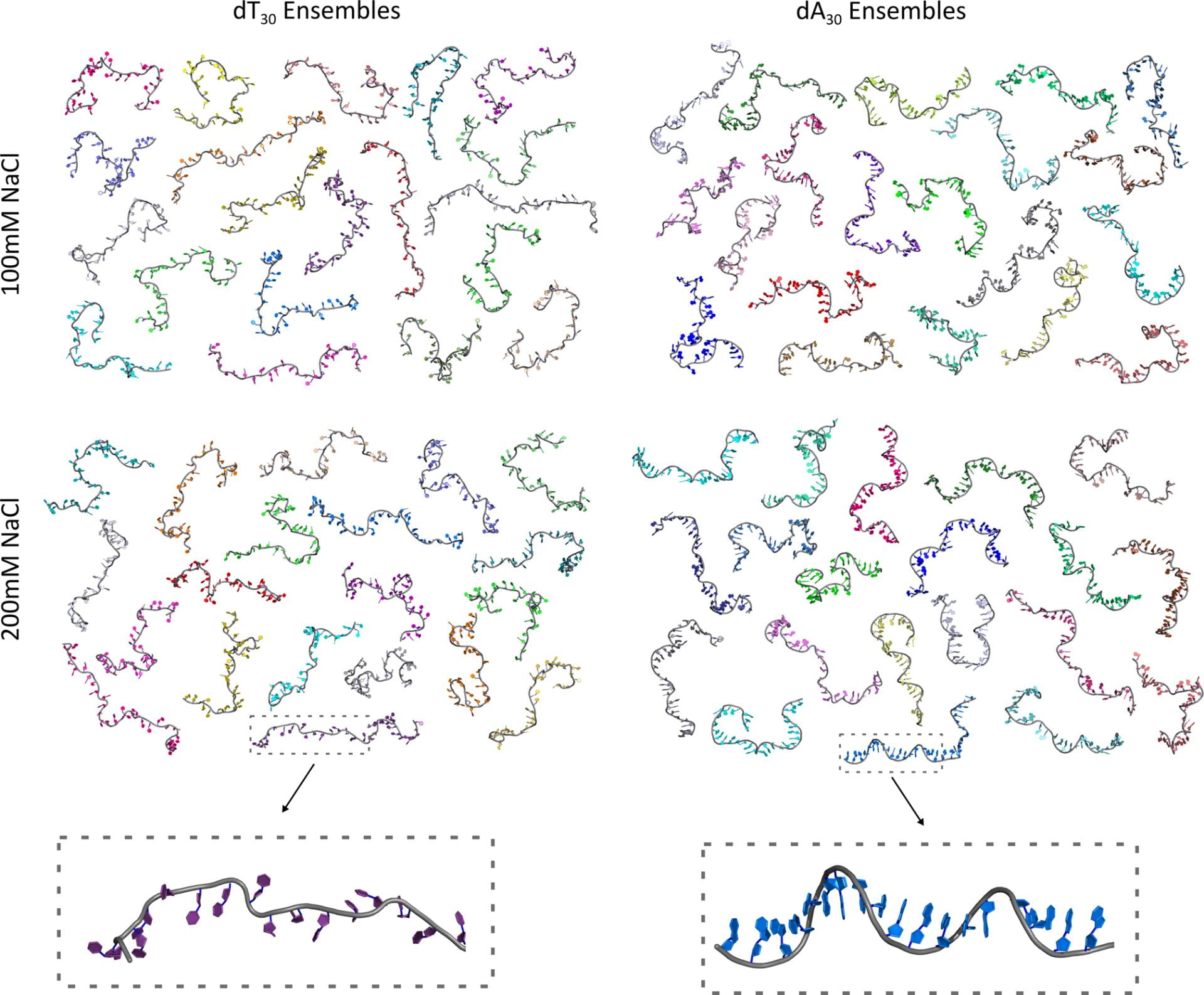
A. Plumridge, K. Andresen, L. Pollack
Visualizing disordered single-stranded RNA: connecting sequence, structure, and electrostatics
J. Am. Chem. Soc. 142(1). 109-119 (2020)
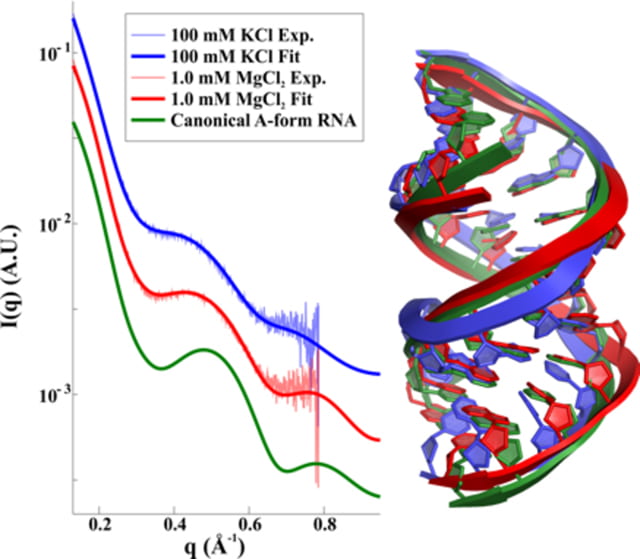
Y. L. Chen, L. Pollack
Salt dependence of A-form RNA duplexes: structures and implications
J Phys. Chem. B. 123(46), 9773-9785 (2019)
G. D. Calvey, A. M. Katz, L. Pollack
Microfluidic Mixing Injector Holder Enables Routine Structural Enzymology Measurements with Mix-and-Inject Serial Crystallography Using X-ray Free Electron Lasers
Analytical Chemistry 91(11), 7139-7144 (2019)
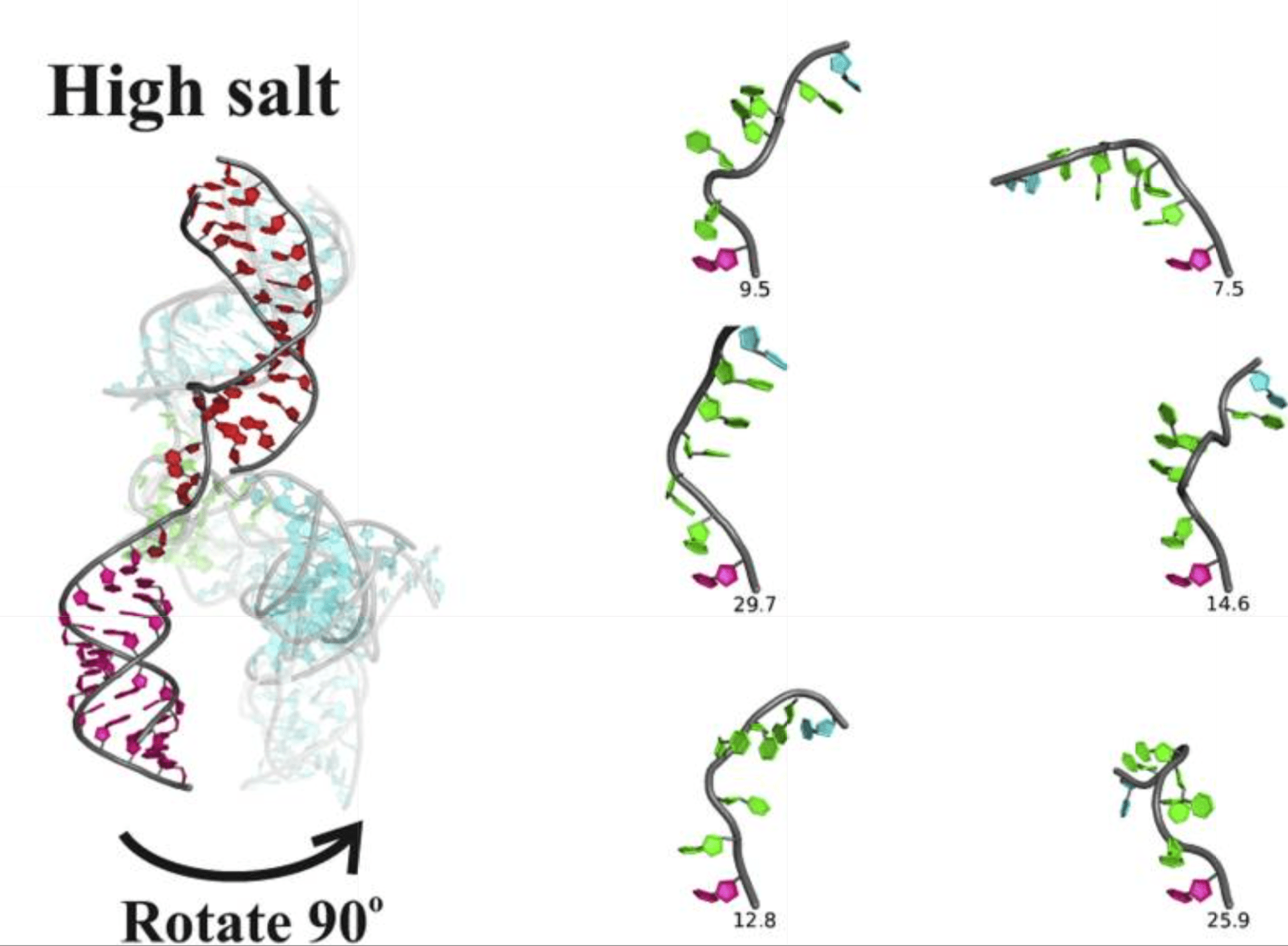
Y. L. Chen, T. Lee, R. Elber, L. Pollack
Conformations of an RNA Helix-Junction-Helix Construct Revealed by SAXS Refinement of MD Simulations
Biophys. J. 116(1), 19-30 (2019)
R. Welty, S. A. Pabit, A. M. Katz, G. D. Calvey, L. Pollack, K. B. Hall
Divalent ions tune the kinetics of a bacterial GTPase center rRNA folding transition from secondary to tertiary structure
RNA 24(12), 1828-1838 (2018)
J. L. Olmos, S. Pandey, J. M. Martin-Garcia, G. Calvey, A. Katz, J. Knoska, C. Kupitz, M. S. Hunter, M. Liang, D. Oberthuer, O. Yefanov, M. Wiedorn, M. Heyman, M. Holl, K. Pande, A. Barty, M. D. Miller, S. Stern, S. Roy-Chowdhury, J. Coe, N. Nagaratnam, J. Zook, J. Verburgt, T. Norwood, I. Poudyal, D. Xu, J. Koglin, M. H. Seaberg, Y. Zhao, S. Bajt, T. Grant, V. Mariani, G. Nelson, G. Subramanian, E. Bae, R. Fromme, R. Fung, P. Schwander, M. Frank, T. A. White, U. Weierstall, N. Zatsepin, J. Spence, P. Fromme, H. N. Chapman, L. Pollack, L. Tremblay, A. Ourmazd, G. N. Phillips, M. Schmidt
Enzyme intermediates captured “on the fly” by mix-and-inject serial crystallography
BMC biology 16, 59 (2018)
Y. L. Chen, J. L. Sutton, L. Pollack
How the Conformations of an Internal Junction Contribute to Fold an RNA Domain
J. Phys. Chem. B. 122(49), 11363-11372 (2018)
A. W. Mauney, J. M. Tokuda, L. M. Gloss, O. Gonzalez, L. Pollack
Local DNA sequence controls asymmetry of DNA unwrapping from nucleosome core particles
Biophys. J. 115(5), 773-781 (2018)
A. Plumridge, A. M. Katz, G. D. Calvey, R. Elber, S. Kirmizialtin, L. Pollack
Revealing the distinct folding phases of an RNA three-helix junction
Nucleic acids research 46(14), 7354-7365 (2018)
S. P. Meisberger, M. A. Warkentin, J. B. Hopkins, A. M. Katz, L. Pollack, R. E. Thorne
Apparatus and methods for low temperature small angle X-ray scattering
US Patent 9927336 (2018)
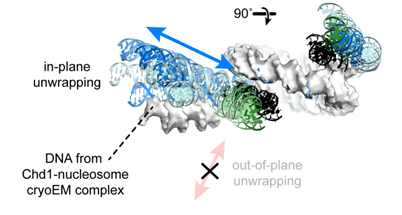
J. M. Tokuda, R. Ren, R. F. Levendosky, R. J. Tay, M. Yan, L. Pollack, G. D. Bowman
The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome
Nucleic acids research 46(10), 4978-4990 (2018)
Y. Sun, A. P. Roznowski, J. M. Tokuda, T. Klose, A. Mauney, L. Pollack, B. A. Fane, M. G. Rossmann
Structural changes of tailless bacteriophage ?X174 during penetration of bacterial cell walls
Proc. Natl. Acad. Sci. USA. 114(52), 13708-13713 (2017)
J. Trewhella, A. P. Duff, D. Durand, F. Gabel, J. M. Guss, W. A. Hendrickson, G. L. Hura, D. A. Jacques, N. M. Kirby, A. H. Kwan, J. Perez, L. Pollack, T. M. Ryan, A. Sali, D. Schneidman-Duhovny, T. Schwede, D. I. Svergun, M. Sugiyama, J. A. Tainer, P. Vachette, J. Westbrook, A. E. Whitten
2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update
Acta Crystallographica D73(9), 710-728
D. Oberthuer, J. Knoška, M. O. Wiedorn, K. R. Beyerlein, D. A. Bushnell, E. G. Kovaleva, M. Heymann, L. Gumprecht, R. A. Kirian, A. Barty, V. Mariani, A. Tolstikova, L. Adriano, S. Awel, M. Barthelmess, K. Dörner, P. L. Xavier, O. Yefanov, D. R. James, G. Nelson, D. Wang, G. Calvey, Y. Chen, A. Schmidt, M. Szczepek, S. Frielingsdorf, O. Lenz, E. Snell, P. J. Robinson, B. Šarler, G. Belšak, M. Ma?ek, F. Wilde, A. Aquila, S. Boutet, M. Liang, M. S. Hunter, P. Scheerer, J. D. Lipscomb, U. Weierstall, R. D. Kornberg, J. C. H. Spence, L. Pollack, H. N. Chapman, S. Bajt
Double-flow focused liquid injector for efficient serial femtosecond crystallography
Scientific reports 7, 44628 (2017)
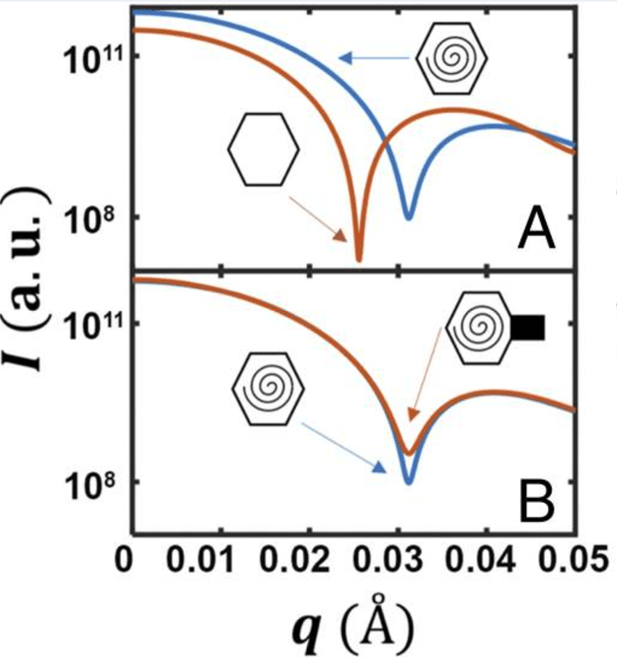
A. Plumridge, S. P. Meisburger, K. Andresen, L. Pollack.
The impact of base stacking on the conformations and electrostatics of single-stranded DNA.
Nucleic Acids Research 45(7), 3932–3943 (2017)
C. Kupitz, J. L. Olmos Jr, M. Holl, L. Tremblay, K. Pande, S. Pandey, D. Oberthür, M. Hunter, M. Liang, A. Aquila, J. Tenboer, G. Calvey, A. M. Katz, Y. Chen, M. O. Wiedorn, J. Knoska, A. Meents, V. Majriani, T. Norwood, I. Poudyal, T. Grant, M. D. Miller, W. Xu, A Tolstikova, A. Morgan, M. Metz, J. M. Martin-Garcia, J. D. Zook, S. Roy-Chowdhury, J. Coe, N. Nagaratnam, D. Meza, R. Fromme, S. Basu, M. Frank, T. White, A. Barty, S. Bajt, O. Yefanov, H. N. Chapman, N. Zatsepin, G. Nelson, U. Weierstall, J. Spence, P. Schwander, L. Pollack, P. Fromme, A. Ourmazd, G. N. Phillips Jr, M. Schmidt.
Structural enzymology using X-ray free electron lasers.
Struct. Dyn. 4(4), 044003 (2017)
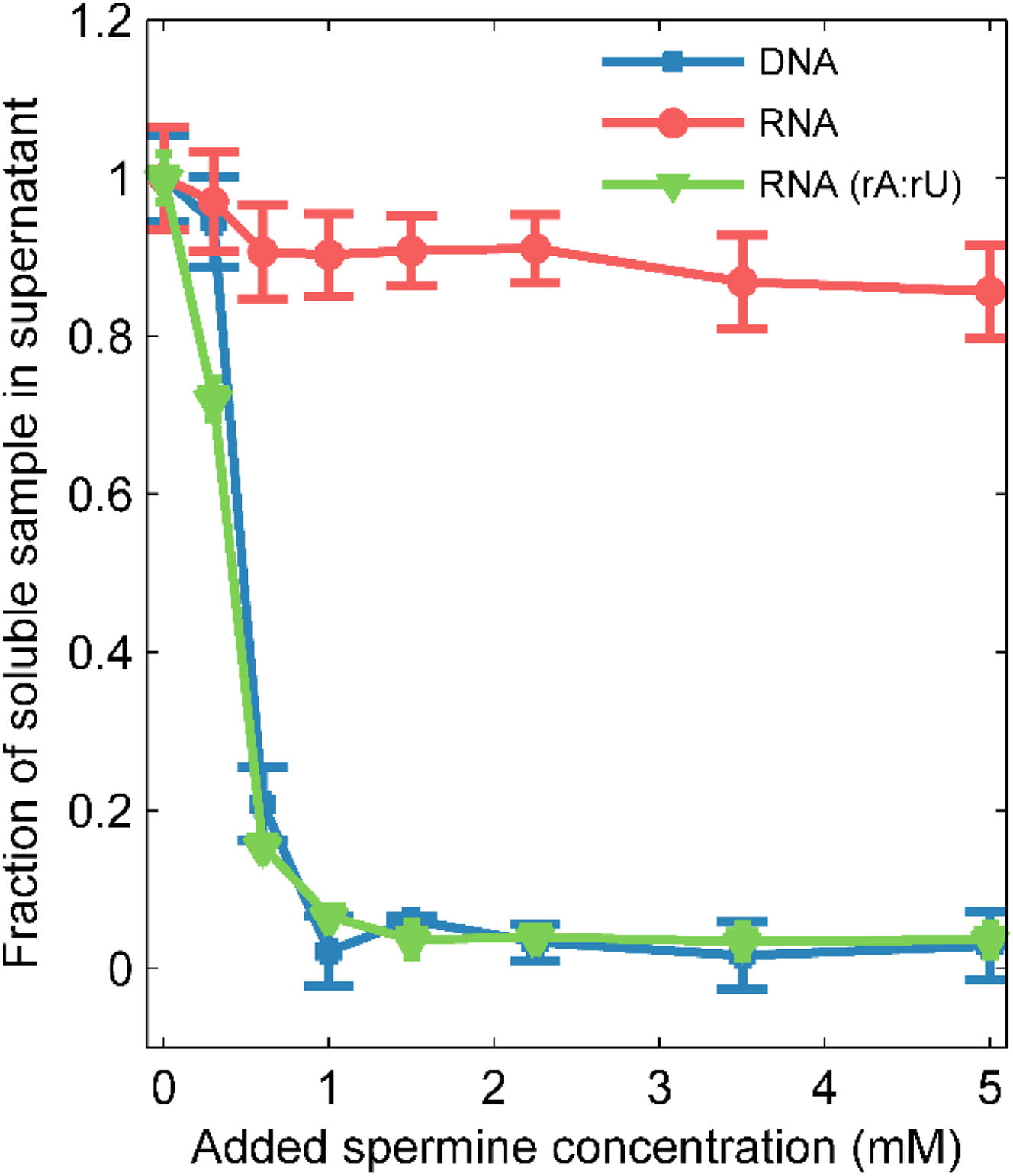
A. M. Katz, I. S. Tolokh, S. A. Pabit, A. V. Onufriev, L. Pollack
Spermine Condenses DNA, but Not RNA Duplexes.
Biophys. J. 112(1), 22-30 (2017)
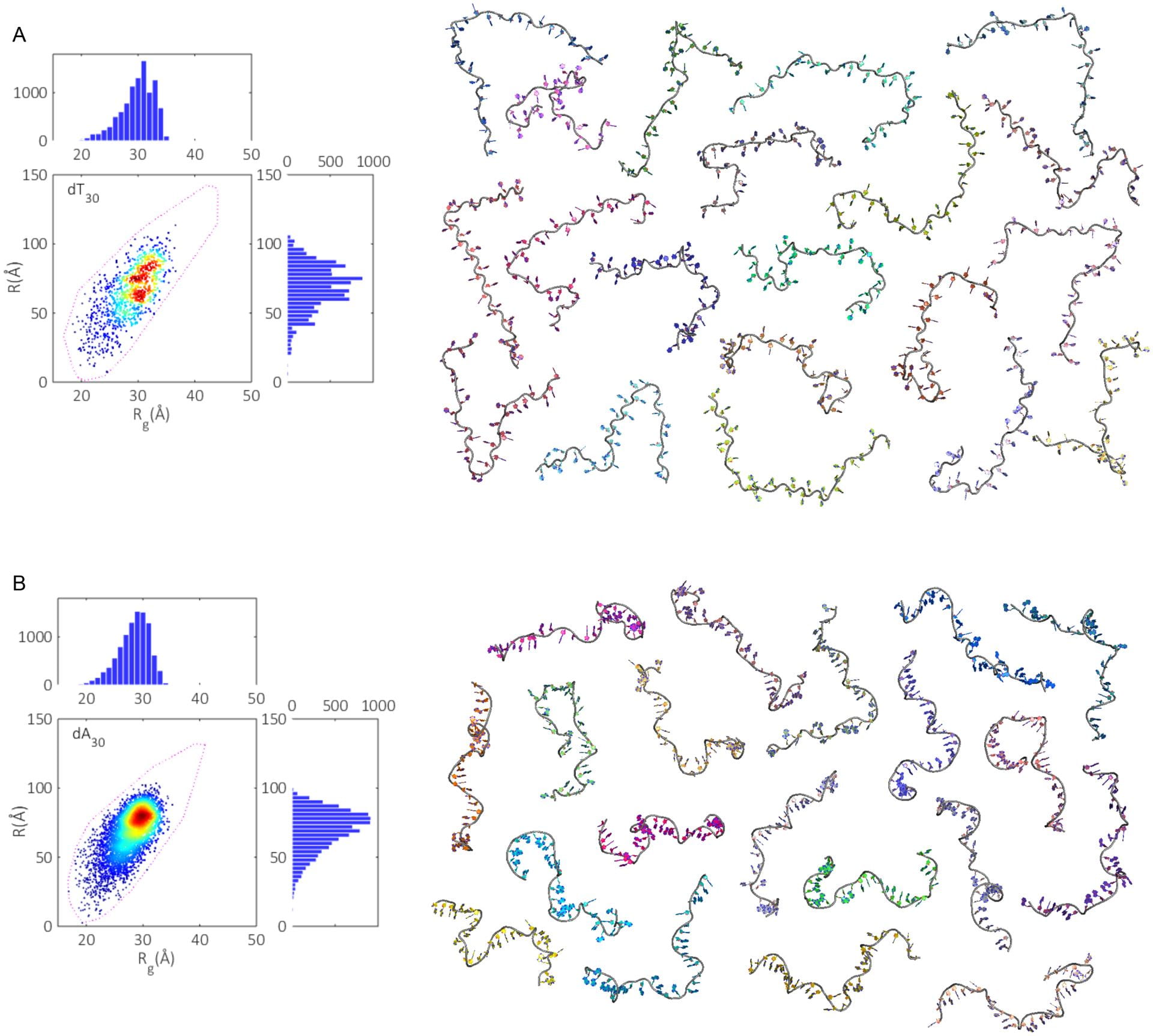
A. Plumridge, S. P. Meisburger, L. Pollack
Visualizing single-stranded nucleic acids in solution.
Nucleic Acids Research. 45(9), 66 (2016)
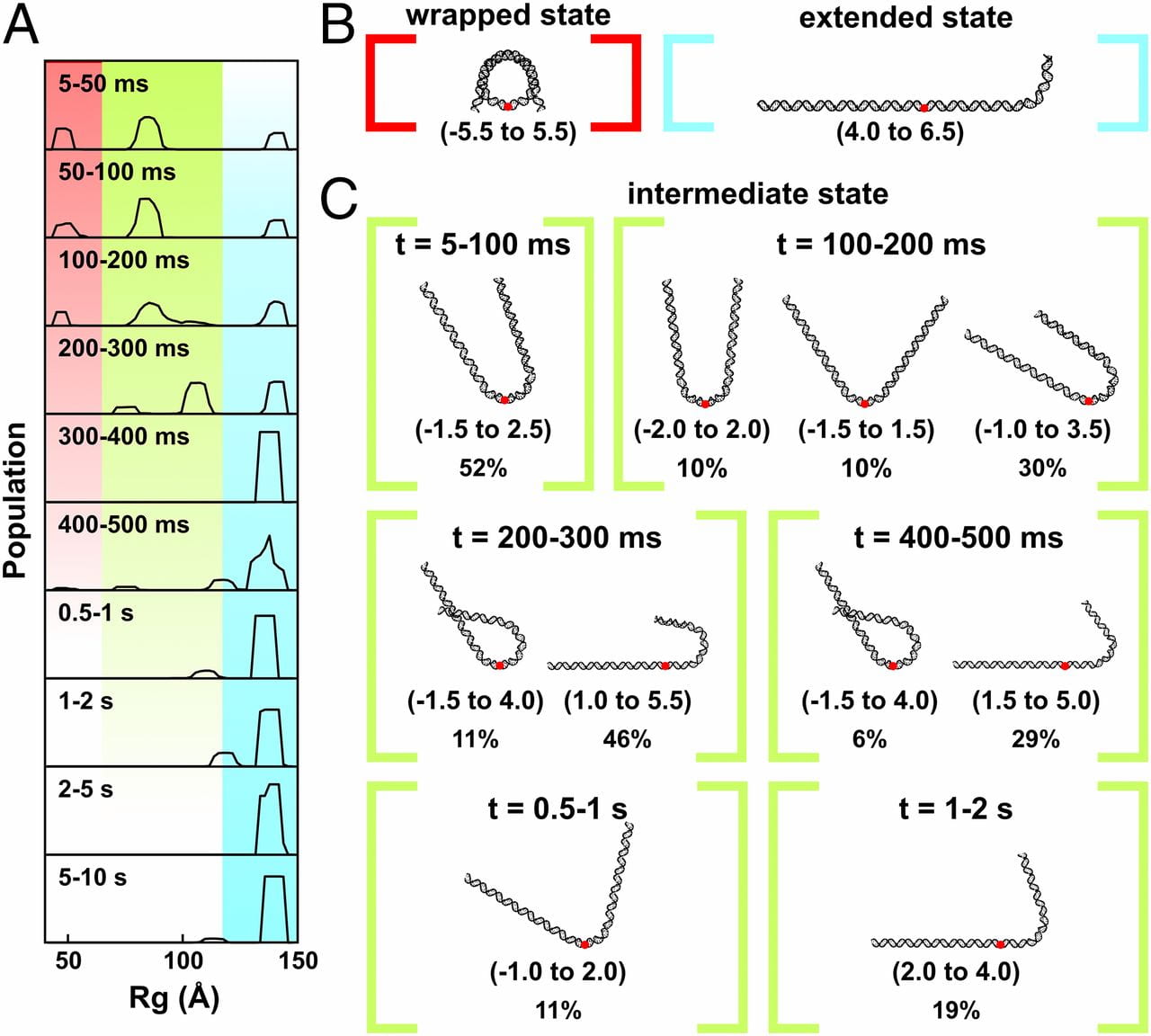
Y. Chen, J. M. Tokuda, T. Topping, S. P. Meisburger, S. A. Pabit, L. M. Gloss, L. Pollack
Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core.
Proc. Natl. Acad. Sci. USA. 114(2), 334-339 (2016)
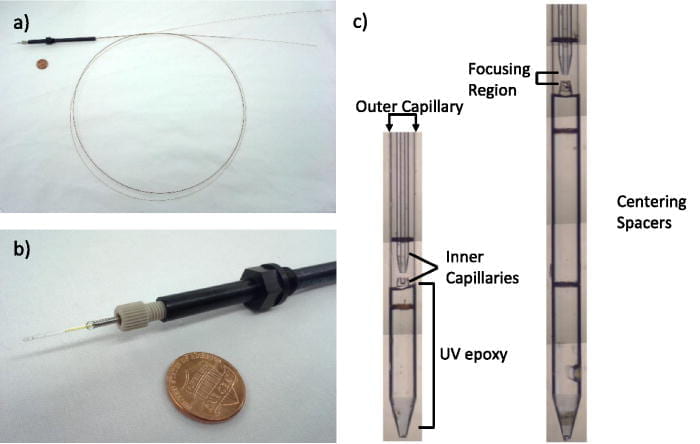
G. D. Calvey, A. M. Katz, C. B. Schaffer, L. Pollack
Mixing injector enables time-resolved crystallography with high hit rate at X-ray free electron lasers.
Struct. Dyn. 3(5), 054301 (2016)

M. Yan, S. Chakravarthy, J. M. Tokuda, L. Pollack, G. D. Bowman, Y.-S. Lee
SAICAR activates PKM2 in its dimeric form.
Biochemistry. 55(33), 4731-4736 (2016)
A. V. Drozdetski, I. S. Tolokh, L. Pollack, N. A. Baker, A. V. Onufriev
Opposing Effects of Multivalent Ions on the Flexibility of DNA and RNA.
Phys. Rev. Lett. 117(2), 028101 (2016)
I. S. Tolokh, A. V. Drozdetski, L. Pollack, N. A. Baker, A. V. Onufriev
Multi-shell model of ion-induced nucleic acid condensation.
J. Chem. Phys. 144(15), 115101 (2016)
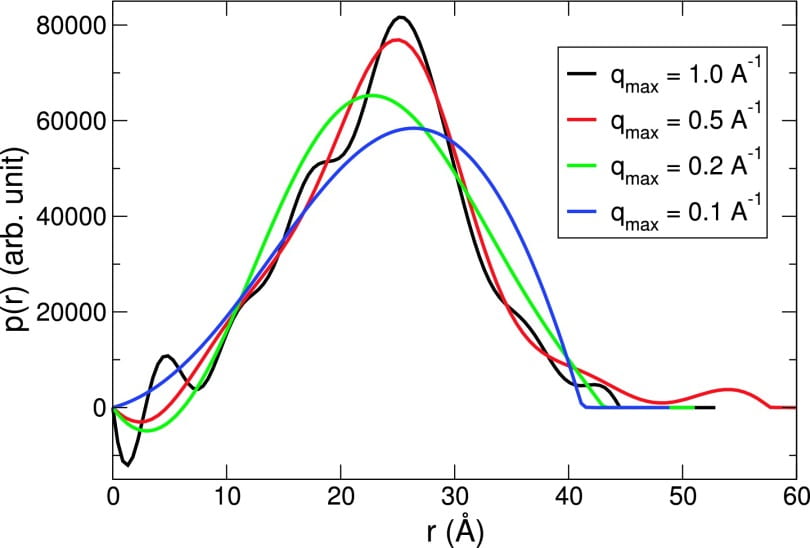
H. T. Nguyen, S. A. Pabit, L. Pollack, D. A. Case
Extracting water and ion distributions from solution x-ray scattering experiments.
J. Chem. Phys. 144(21). 214105 (2016)
S. A. Pabit, A. M. Katz, I. S. Tolokh, A. Drozdetski, N. Baker, A. V. Onufriev, L. Pollack
Understanding nucleic acid structural changes by comparing wide-angle x-ray scattering (WAXS) experiments to molecular dynamics simulations.
J. Chem. Phys. 144(20), 205102 (2016)
Y. Chen, L. Pollack
SAXS studies of RNA: structures, dynamics, and interactions with partners.
Wiley Interdisp Rev RNA. 7(4), 512-526 (2016)
J. M. Tokuda, S. A. Pabit, L. Pollack
Protein-DNA and ion-DNA interactions revealed through contrast variation SAXS.
Biophys. Rev. 8(2), 139-149 (2016)
M. L. Sushko, D. G. Thomas, S. A. Pabit, L. Pollack, A. V. Onufriev, N. A. Baker
The Role of Correlation and Solvation in Ion Interactions with B-DNA.
Biophys. J. 110(2), 315-26 (2016)
J. L. Sutton, L. Pollack
Tuning RNA Flexibility with Helix Length and Junction Sequence.
Biophys. J. 109(12), 2644-53 (2015)
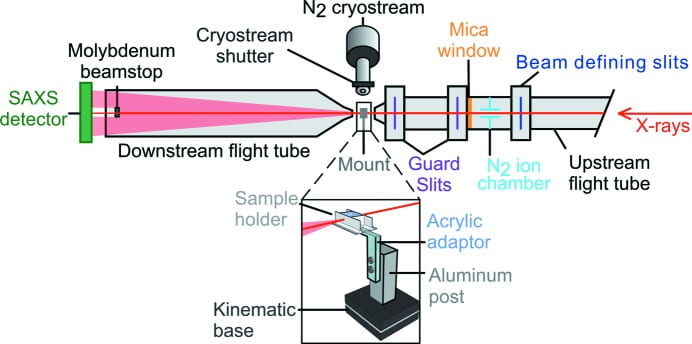
J. B. Hopkins, A. M. Katz, S. P. Meisburger, M. A. Warkentin, R. E. Thorne, L. Pollack
A microfabricated fixed path length silicon sample holder improves background subtraction for cryoSAXS.
J. Appl. Cryst. 48, 227-237 (2015)
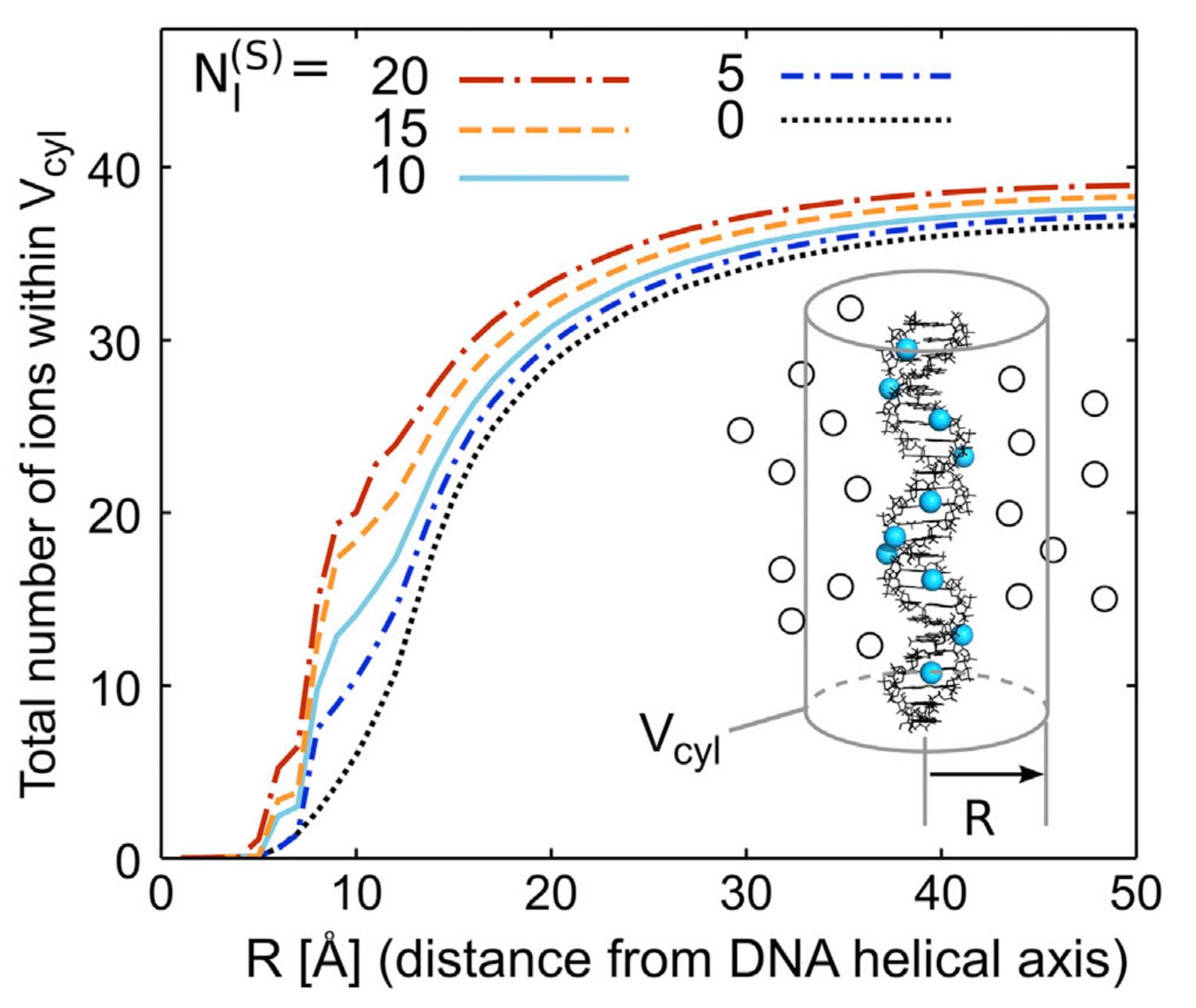
S. P. Meisburger, S. A. Pabit, L. Pollack
Determining the Locations of Ions and Water around DNA from X-Ray Scattering Measurements.
Biophys. J. 108(12), 2886-95 (2015)
D. Mizrachi, Y. Chen, J. Liu, H. M. Peng, A. Ke, L. Pollack, R. J. Turner, R. J. Auchus, M. P. DeLisa
Making water-soluble integral membrane proteins in vivo using an amphipathic protein fusion strategy.
Nat. Commun. 6, 6826 (2015)
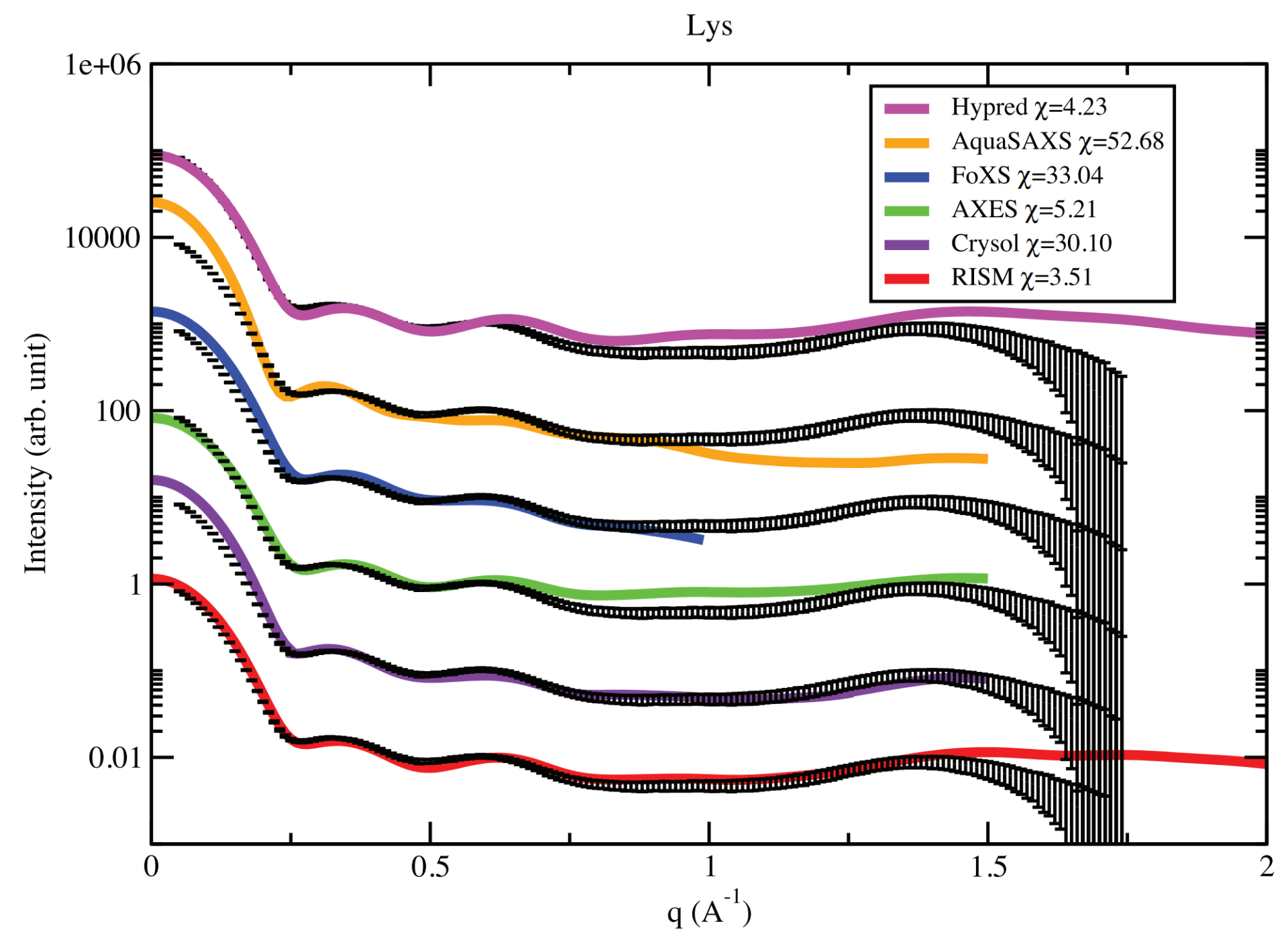
H. T. Nguyen, S. A. Pabit, S. P. Meisburger, L. Pollack, D. A. Case
Accurate small and wide angle x-ray scattering profiles from atomic models of proteins and nucleic acids.
J. Chem. Phys. 141(22) (2014)
D. Wang, U. Weierstall, L. Pollack, J. Spence
Double-focusing mixing jet for XFEL study of chemical kinetics.
J. Synch. Rad. 21(6), 1364-6 (2014)
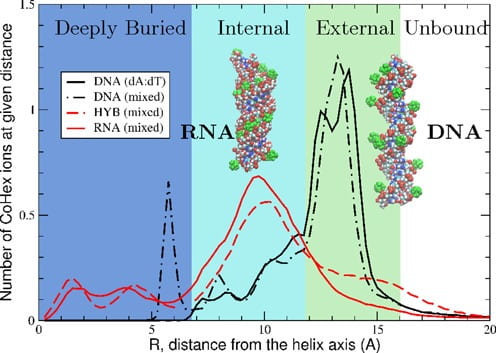
I. S. Tolokh, S. A. Pabit, A. M. Katz, Y. Chen, A. Drozdetski, N. Baker, L. Pollack, A. V. Onufriev
Why double-stranded RNA resists condensation.
Nucleic Acids Research 42(16), 10823-31 (2014)
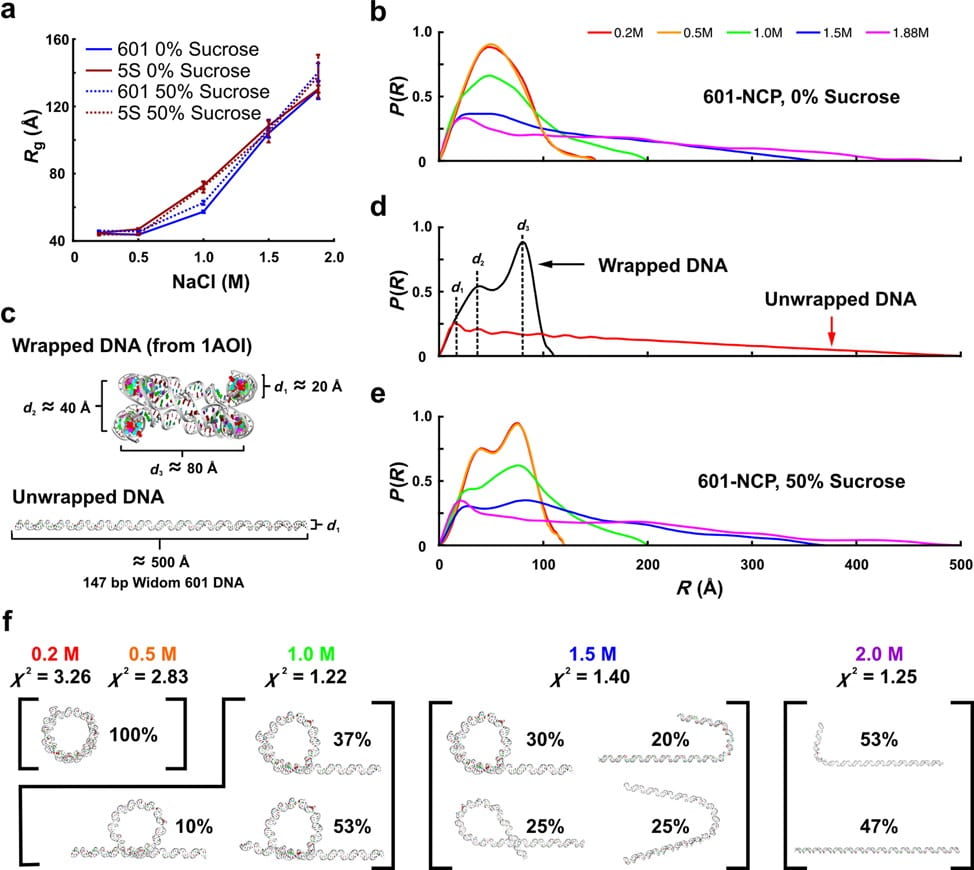
Y. Chen, J. M. Tokuda, T. Topping, J. L. Sutton, S.P. Meisburger, S. A. Pabit, L. M. Gloss, L. Pollack
Revealing transient structures of nucleosomes as DNA unwinds.
Nucleic Acids Research 42(13), 8767-76 (2014)
A. T. Vaidya, D. Top, C. C. Manahan, J. M. Tokuda, S. Zhang, L. Pollack, M. W. Young, B. R. Crane
Flavin reduction activates Drosophila cryptochrome.
Proc. Natl. Acad. Sci. USA 110(51), 20455-60 (2013)
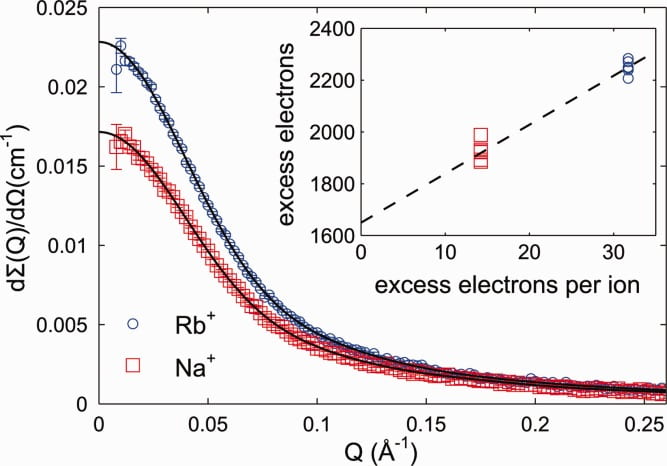
S. P. Meisburger, J. L. Sutton, H. Chen, S.A. Pabit, S. Kirmizialtin, R. Elber, L. Pollack
Polyelectrolyte properties of single stranded DNA measured using SAXS and single-molecule FRET: Beyond the wormlike chain model.
Biopolymers. 99(12), 1032-1045 (2013)
J. C. Grigg, Y. Chen, F. J. Grundy, T. M. Henkin, L. Pollack, A. Ke
T box RNA decodes both the information content and geometry of tRNA to affect gene expression.
Proc. Natl. Acad. Sci. USA 110(18), 7240-5 (2013)
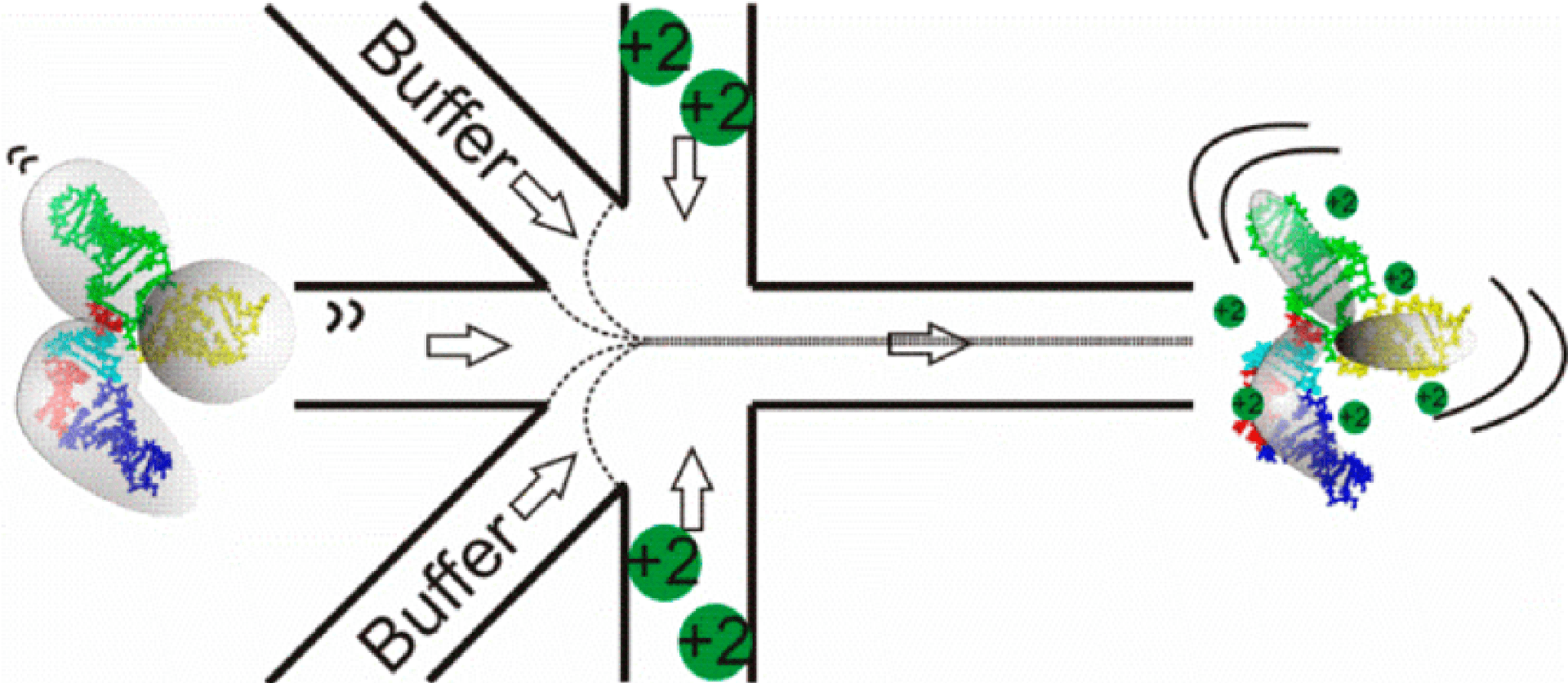
S. A. Pabit, J. L. Sutton, H. Chen, L. Pollack
Role of ion valence in the submillisecond collapse and folding of a small RNA domain.
Biochemistry. 52(9), 1539-46 (2013)
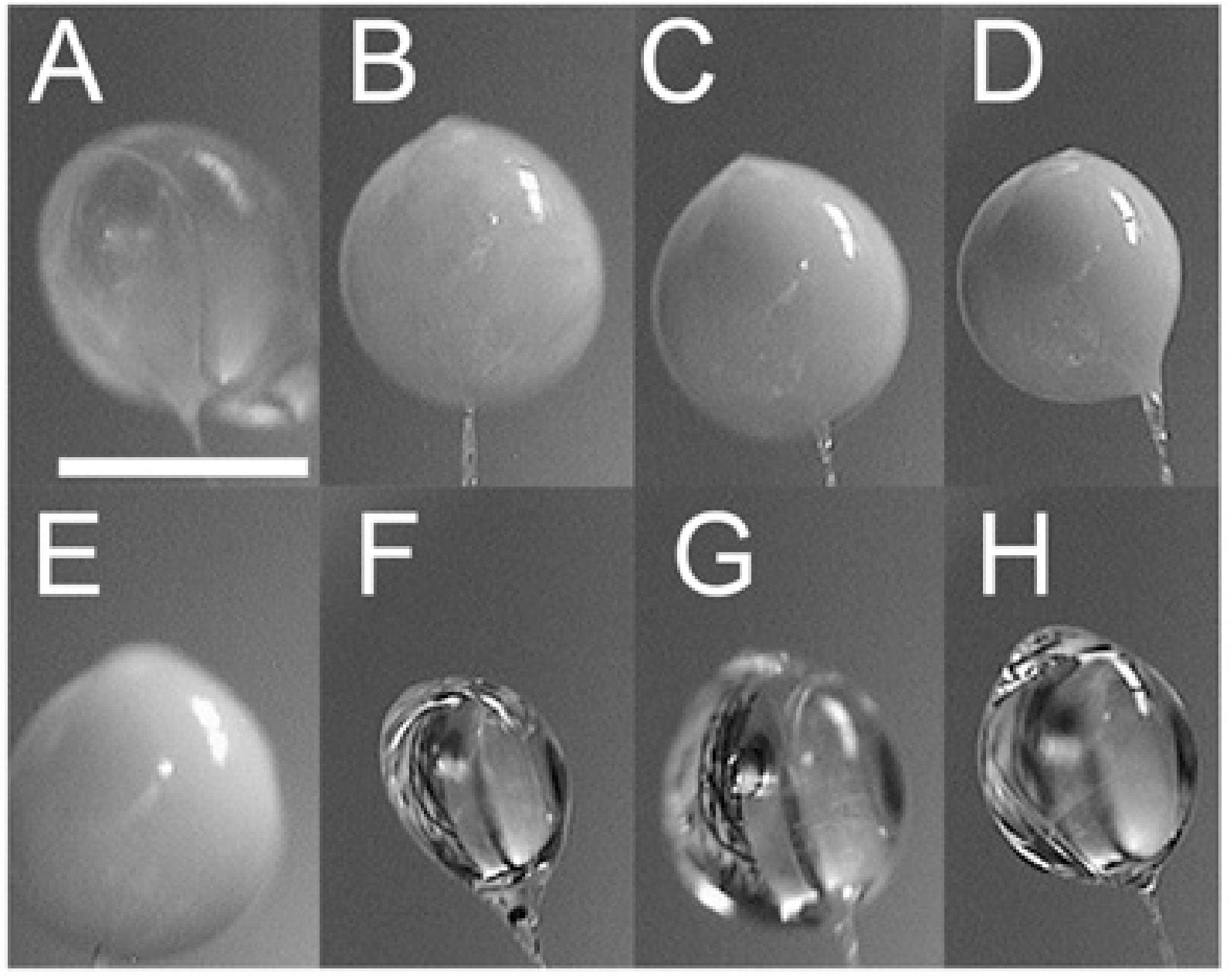
S. P. Meisberger, M. Warkentin, H. Chen, J. B. Hopkins, R. E. Gillilan, L. Pollack, R. E. Thorne
Breaking the radiation damage limit with Cryo-SAXS
Biophys. J. 104(1), 227-36 (2012)
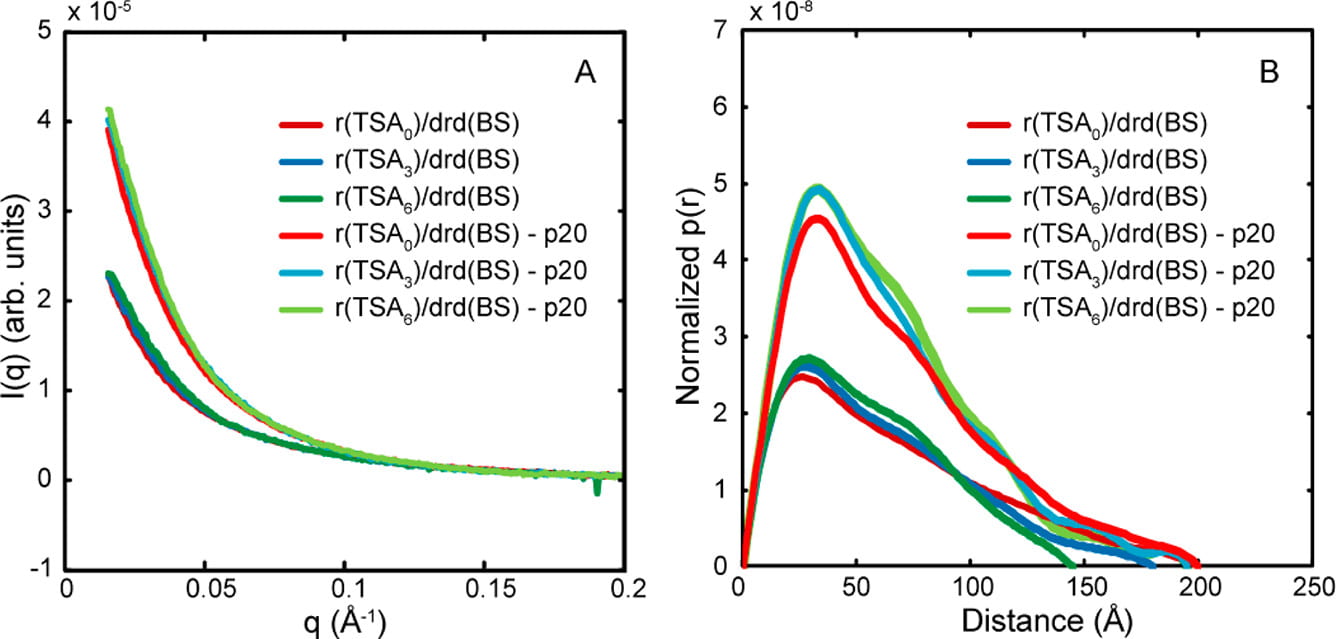
S. Patel, J. M. Blose, J. E. Solowski, L. Pollack, P. C. Bevilacqua
Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering.
Biochemistry. 51(46), 9312-22 (2012)
D. A. Costello, D. W. Lee, J. Drewes, K. A. Vasquez, K. Kisler, U. Weisner, L. Pollack, G. R. Whittaker, S. Daniel
Influenza virus-membrane fusion triggered by proton uncaging for single particle studies of fusion kinetics.
Anal. Chem. 84(20), 8480-9 (2012)
J. C. Schlatterer, M. S. Wieder, C. D. Jones, L. Pollack, M. Brenowitz
Pyrite footprinting of RNA.
Biochem. Biophys. Res. Commun. 425(2), 374-8 (2012)
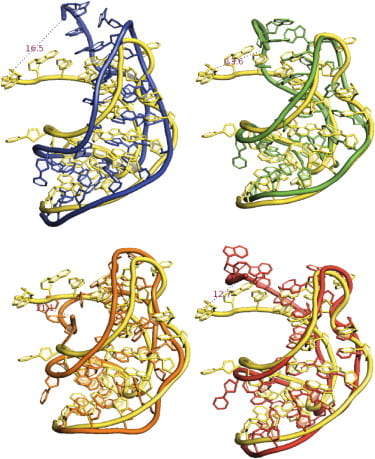
S. Kirmizialtin, S. A. Pabit, S. P. Meisburger, L. Pollack, R. Elber
RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations.
Biophys. J. 102(4), 819-28 (2012)
T. Y. Yoo, S. P. Meisburger, J. Hinshaw, L. Pollack, G. Haran, T. R. Sosnick, K. Plaxco
Small-angle X-ray scattering and single-molecule FRET spectroscopy produce highly divergent views of the low-denaturant unfolded state.
J. Mol. Biol. 418(3-4), 226-36 (2012)
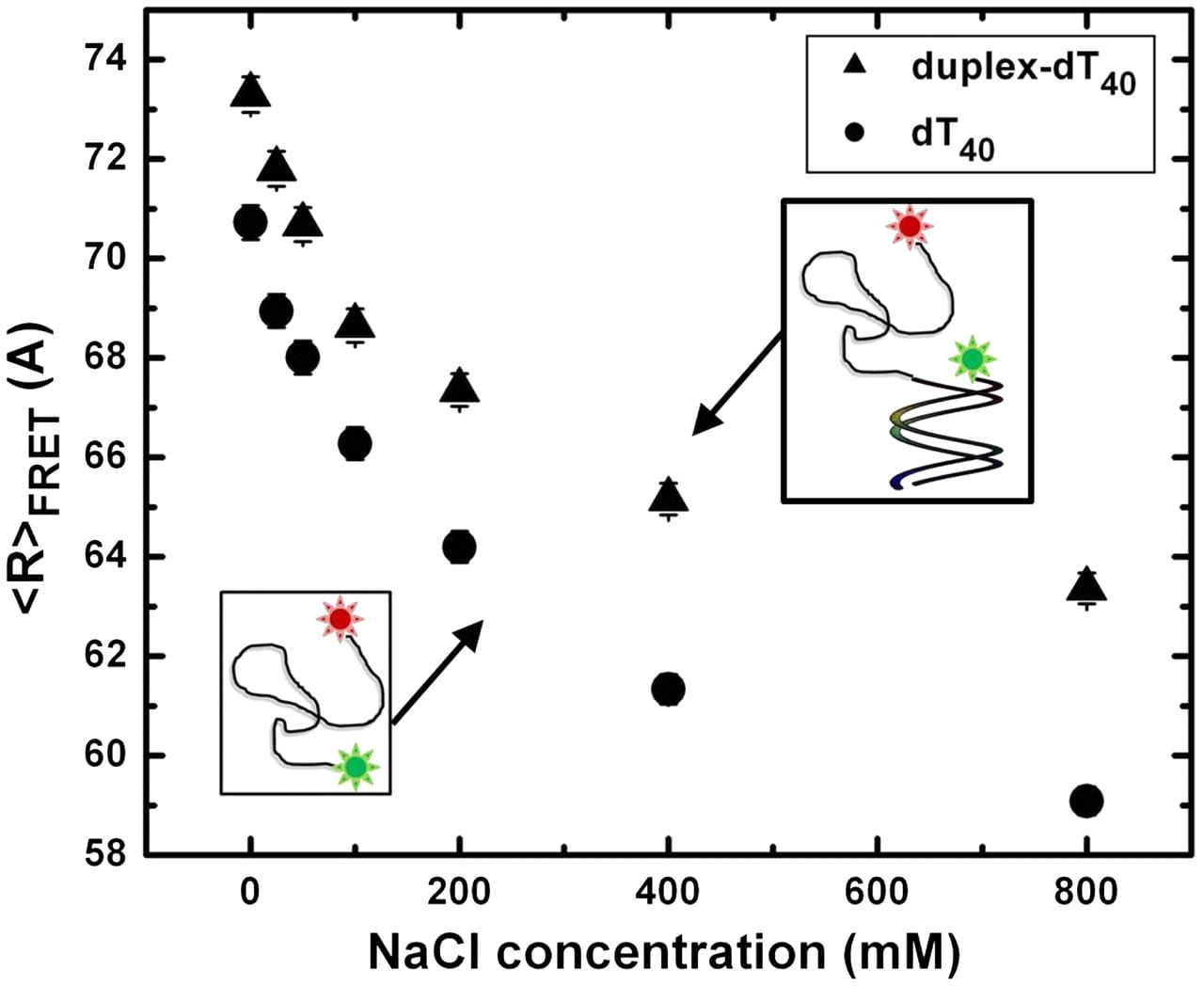
H. Chen, S. P. Meisburger, S. A. Pabit, J. L. Sutton, W. W. Webb, L. Pollack
Ionic strength-dependent persistence lengths of single-stranded RNA and DNA.
Proc. Natl. Acad. Sci. USA 109(3), 799-804 (2012)
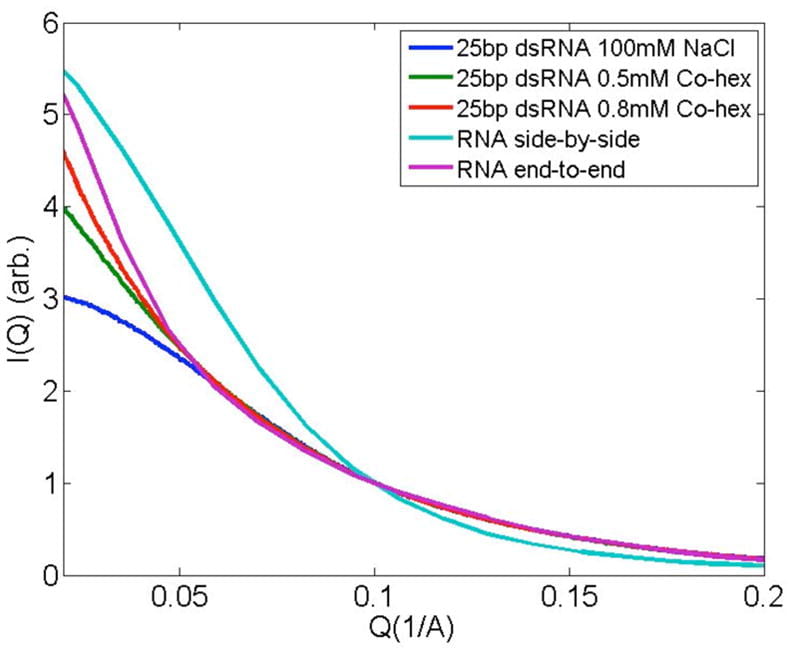
L. Li, S. A. Pabit, S. Meisburger, L. Pollack
Double stranded RNA resists condensation.
Phys. Rev. Lett., 106, 108101 (2011)
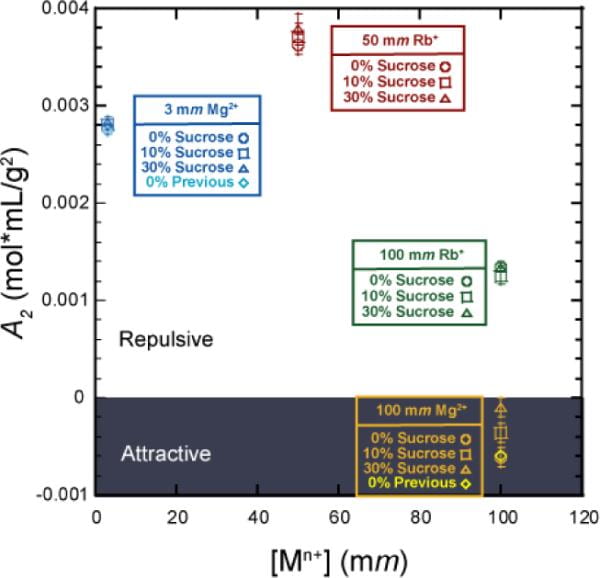
J. M. Blose, S. A. Pabit, S. P. Meisburger, L. Li, C. D. Jones, L. Pollack
Effects of a protecting osmolyte on the ion atmosphere surrounding DNA duplexes.
Biochemistry. 50(40), 8540-7 (2011)
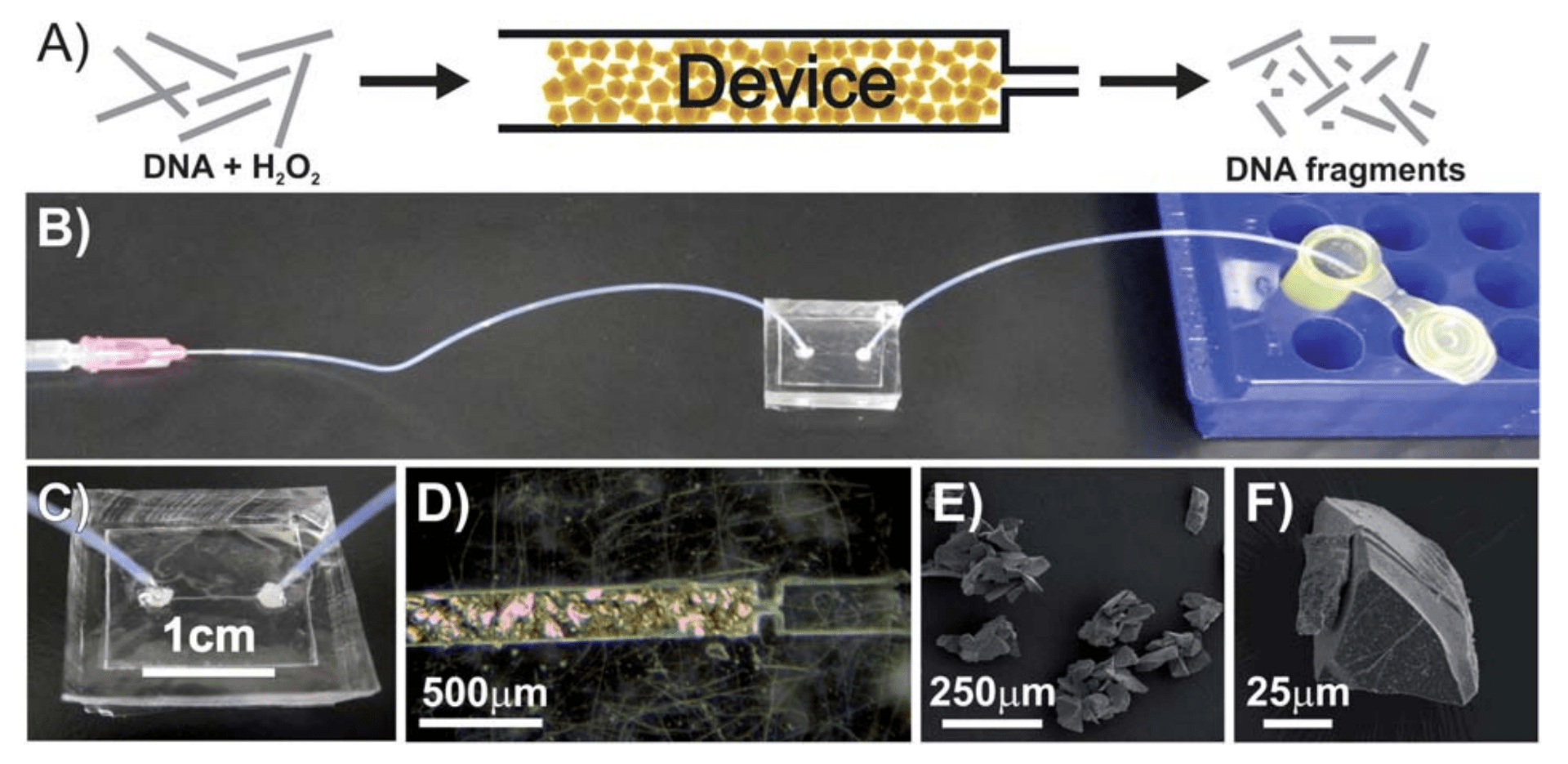
C. D. Jones, J. C. Schlatterer, M. Brenowitz, L. Pollack
A microfluidic device that generates hydroxyl radicals to probe the solvent accessible surface of nucleic acids.
Lab Chip. 11(20), 3458-64 (2011)
L. Pollack
SAXS studies of ion-nucleic acid interactions.
Annu. Rev. Biophys. 40:225-42 (2011)
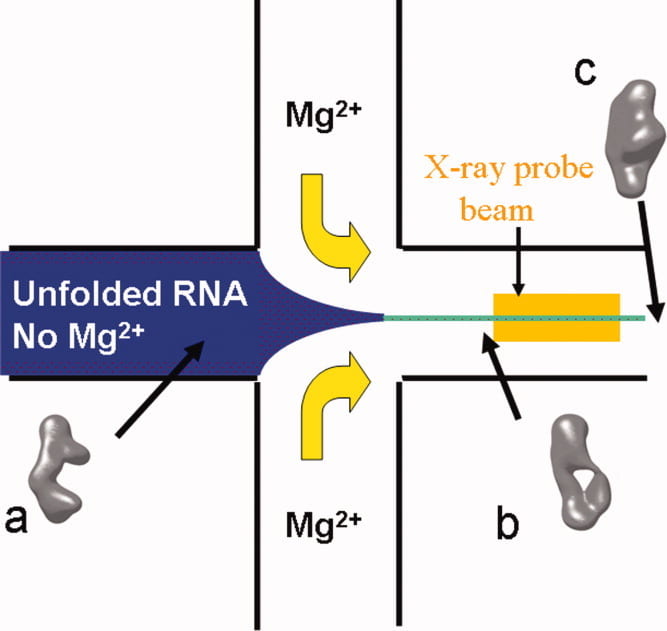
L. Pollack
Time resolved SAXS and RNA folding.
Biopolymers, 95, 54. (2011)
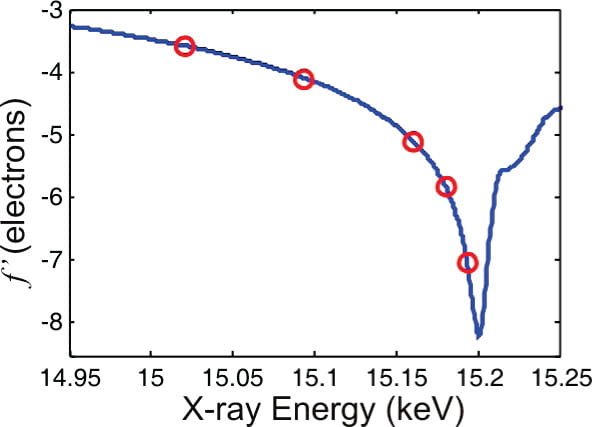
S. A. Pabit, S. P. Meisburger, L. Li, J. M. Blose, C. D. Jones, L. Pollack
Counting ions around DNA with ASAXS.
J. Am. Chem Soc. 132 (46), pp 16336 (2010)
Gerard C.L. Wong, Lois Pollack
Electrostatics of strongly charged biological polymers: Ion-mediated interactions and self-organization in nucleic acids and proteins.
Annual Review of Physical Chemistry Volume 61 (2010)
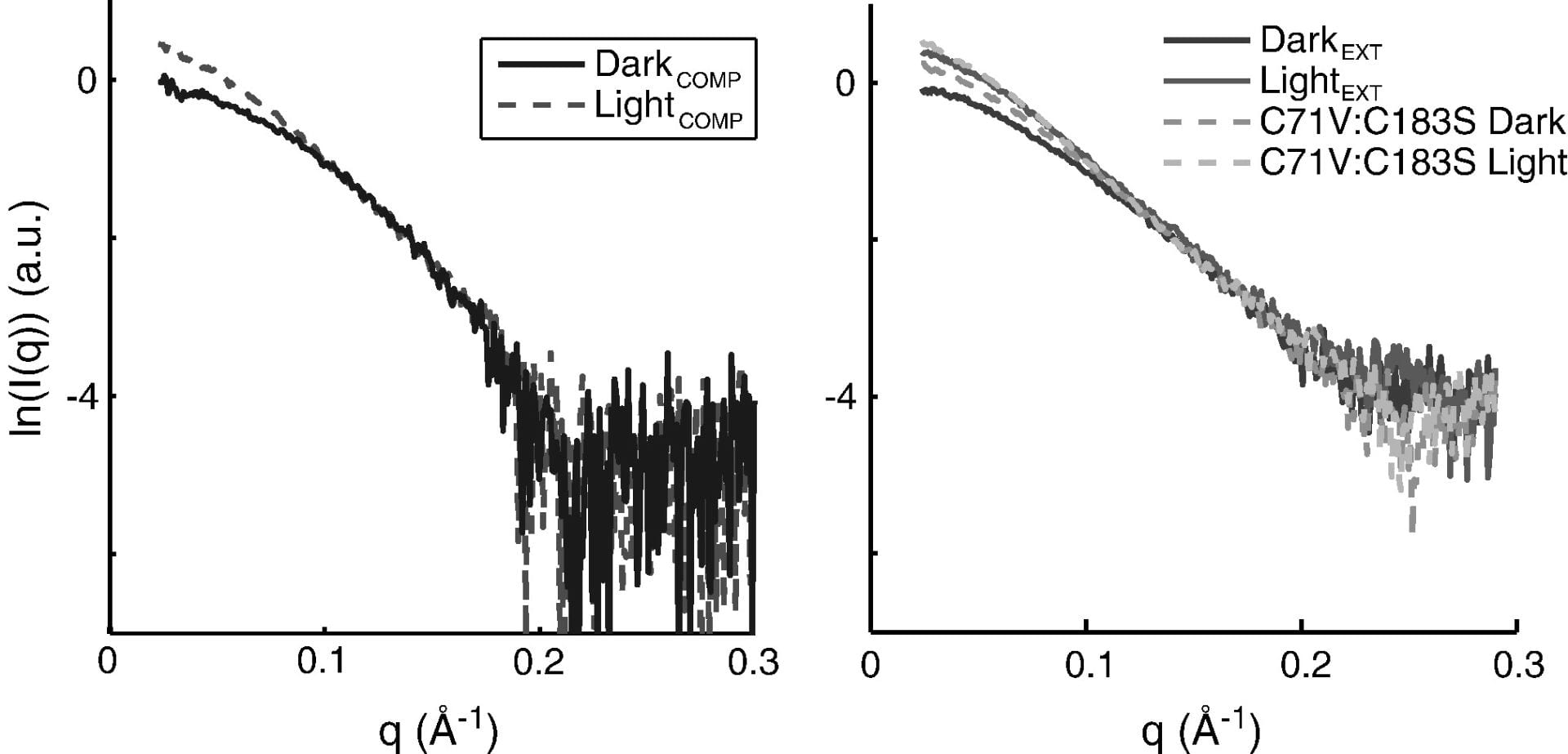
J. S. Lamb, B. Zoltowski, S. Pabit, L. Li, B. Crane, and L. Pollack
Illuminating solution responses of a LOV-domain protein with photocoupled small angle x-ray scattering.
J. Mol. Biol (393), 909-919 (2009)
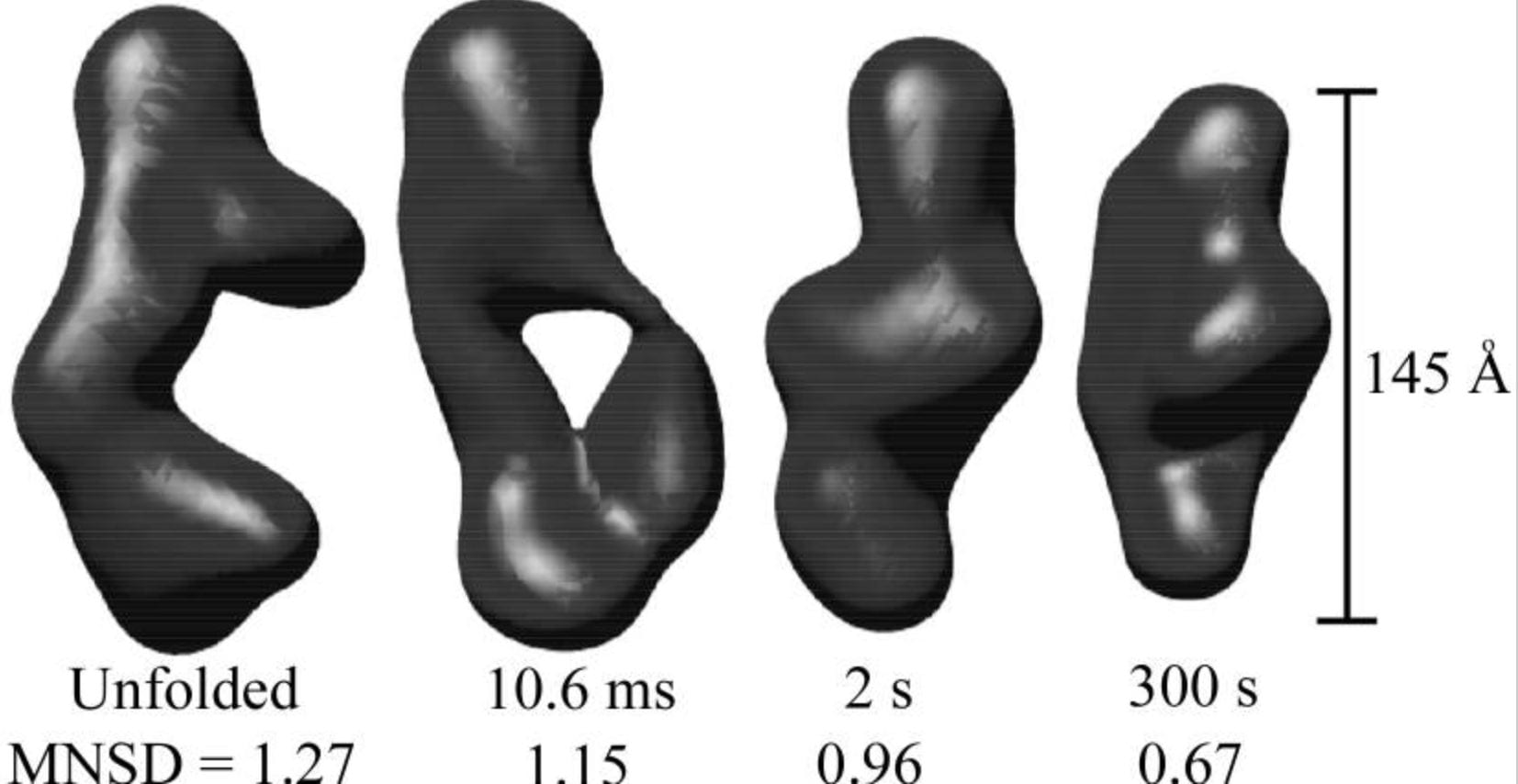
J.S. Lamb, L. Kwok, X. Qiu, K. Andresen, H. Park and L. Pollack
Reconstructing three dimensional shape envelopes from time resolved small angle x-ray scattering data.
J. App. Cryst. 41, 1046 (2008)
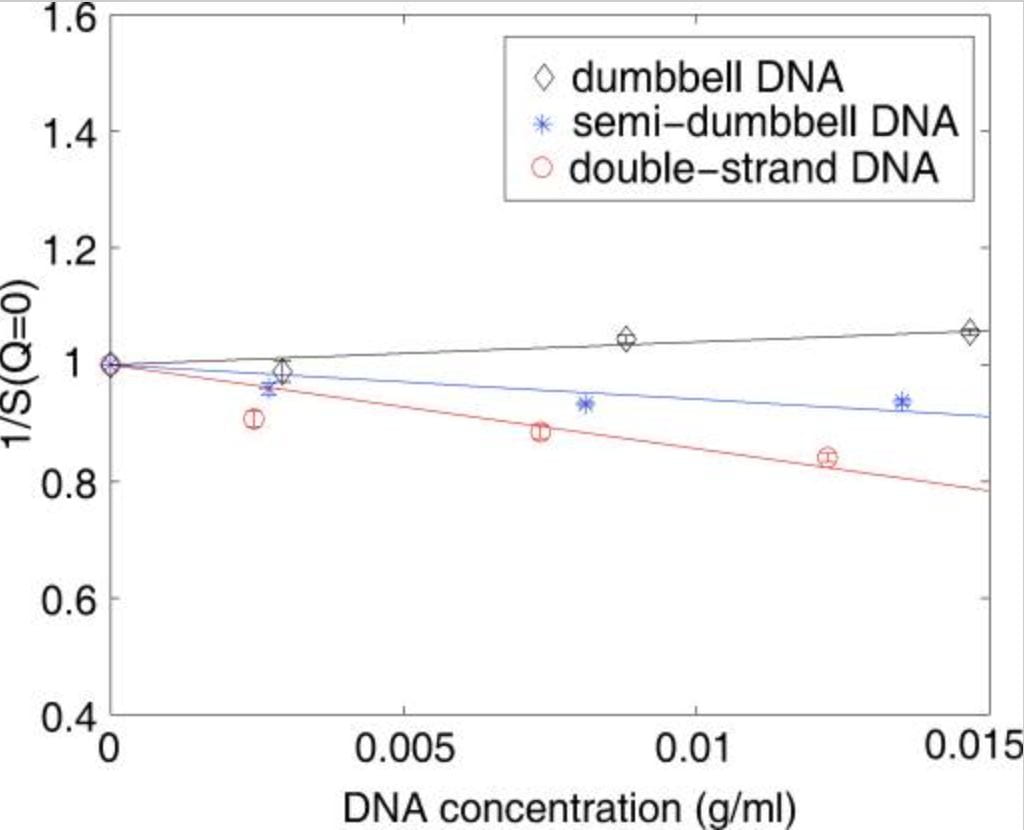
L. Li, S.A. Pabit, J.S. Lamb, H.Y. Park and L. Pollack
Closing the lid on DNA end-to-end stacking interactions.
Appl. Phys. Lett. 92, 223901 (2008)
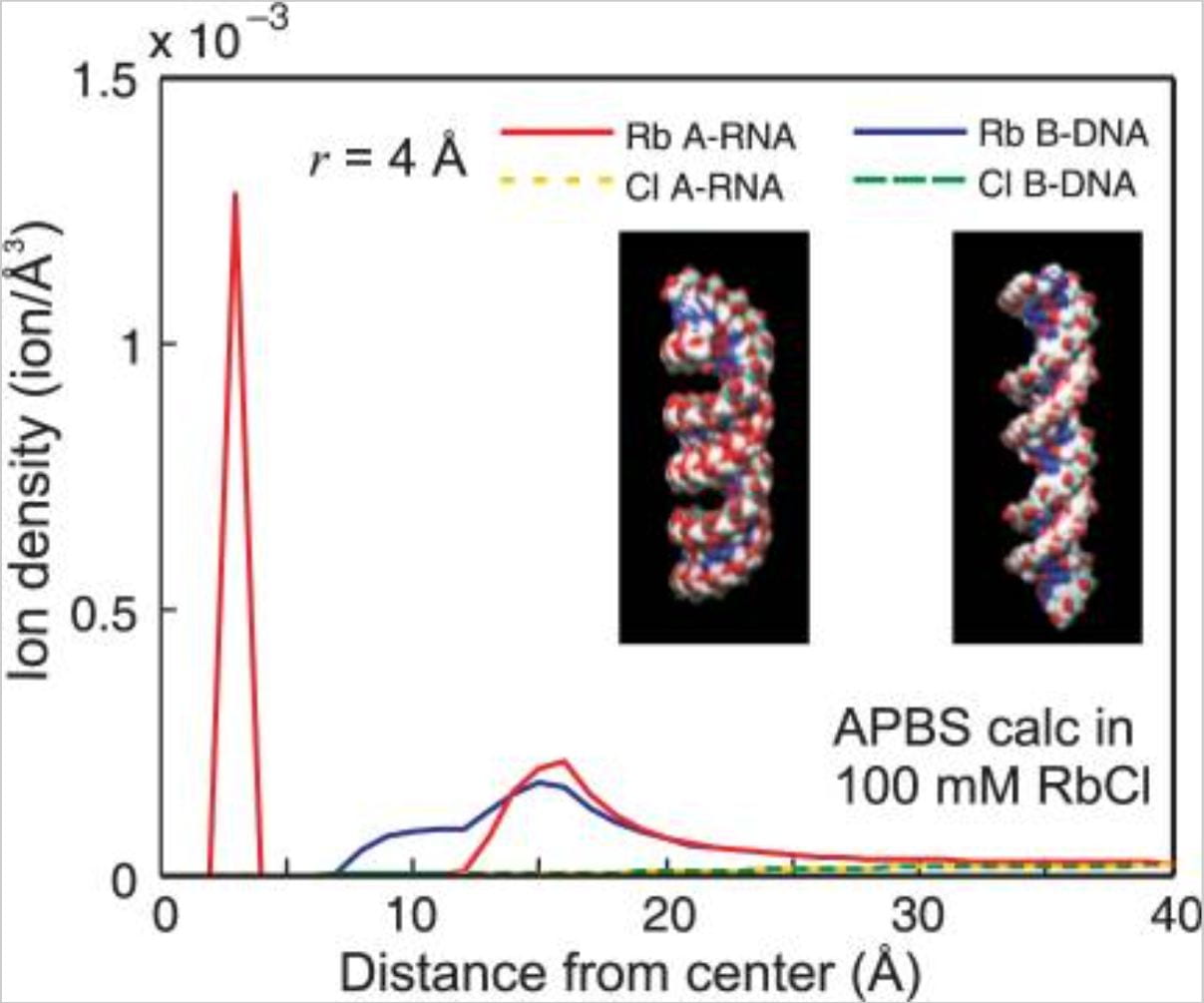
S.A. Pabit, X. Qiu, J.S. Lamb, L. Li, S.P. Meisburger and L. Pollack
Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared to dsDNA.
Nucleic Acids Research 37(12), 3887-96 (2009)
S. A. Pabit, K.D. Finkelstein and L. Pollack
Using anomalous small angle x-ray scattering to probe the ion atmosphere around nucleic acids.
Methods In Enzymology (469), 391-410 (2009)
L. Pollack and S. Doniach
Time resolved x-ray scattering and RNA folding.
Methods In Enzymology (469), 253-268 (2009)
X. Qiu, K. Andresen, J.S. Lamb, L.W. Kwok and L. Pollack
Abrupt transition from free, repulsive to condensed, attractive DNA phase, induced by multivalent polyamine cations.
Phys. Rev. Lett. 101, 228101 (2008)
J.S. Lamb, B. Zoltowski, S. A. Pabit, B. R. Crane and L. Pollack
Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle x-ray scattering.
J. Am. Chem. Soc. 130, 12226 (2008)
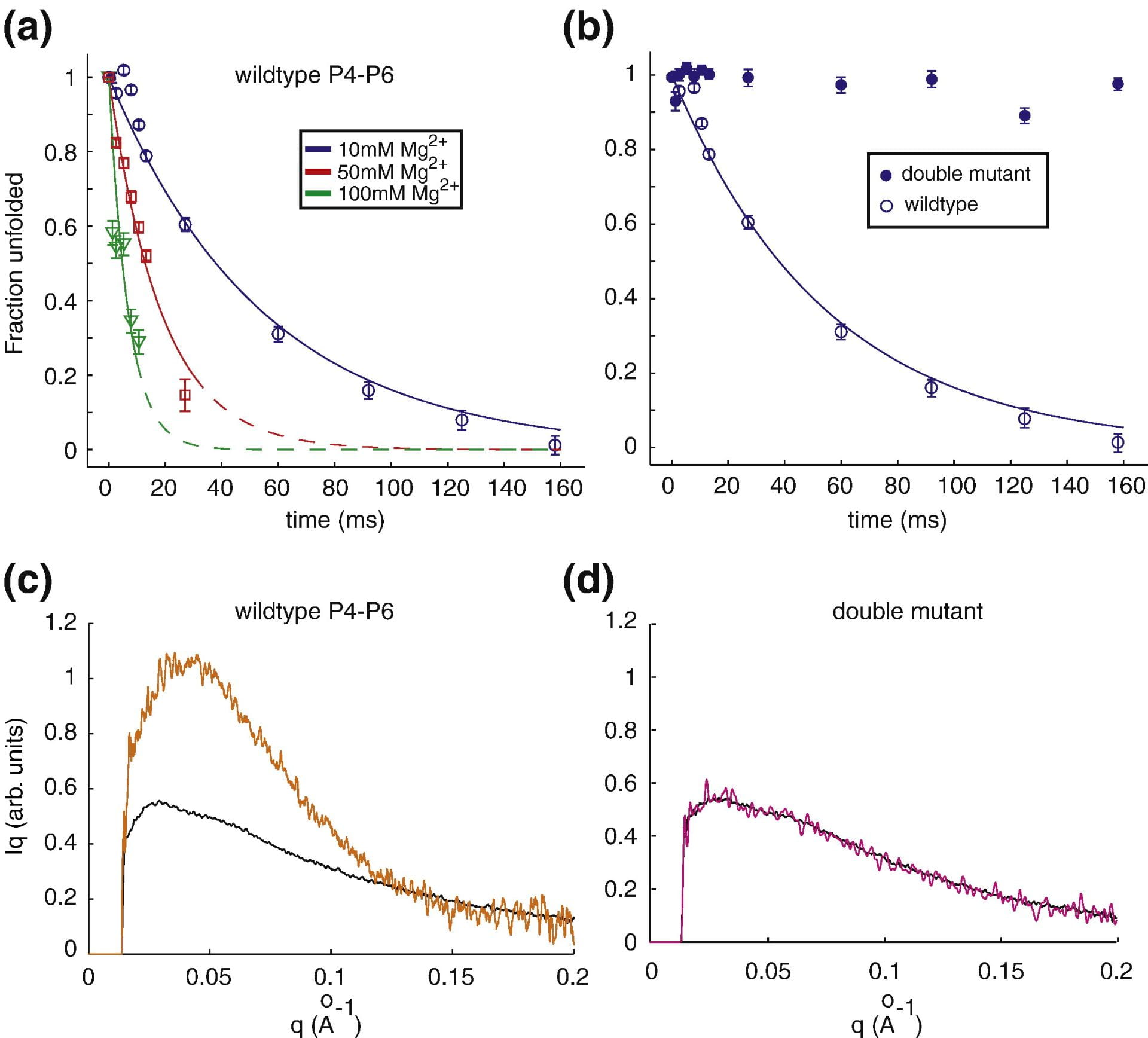
J. Schlatterer, L.W. Kwok, J.S. Lamb, H.Y. Park, K. Andresen, M. Brenowitz and L. Pollack
Hinge stiffness is a barrier to RNA folding.
J. Mol. Biol. 379, 859 (2008)
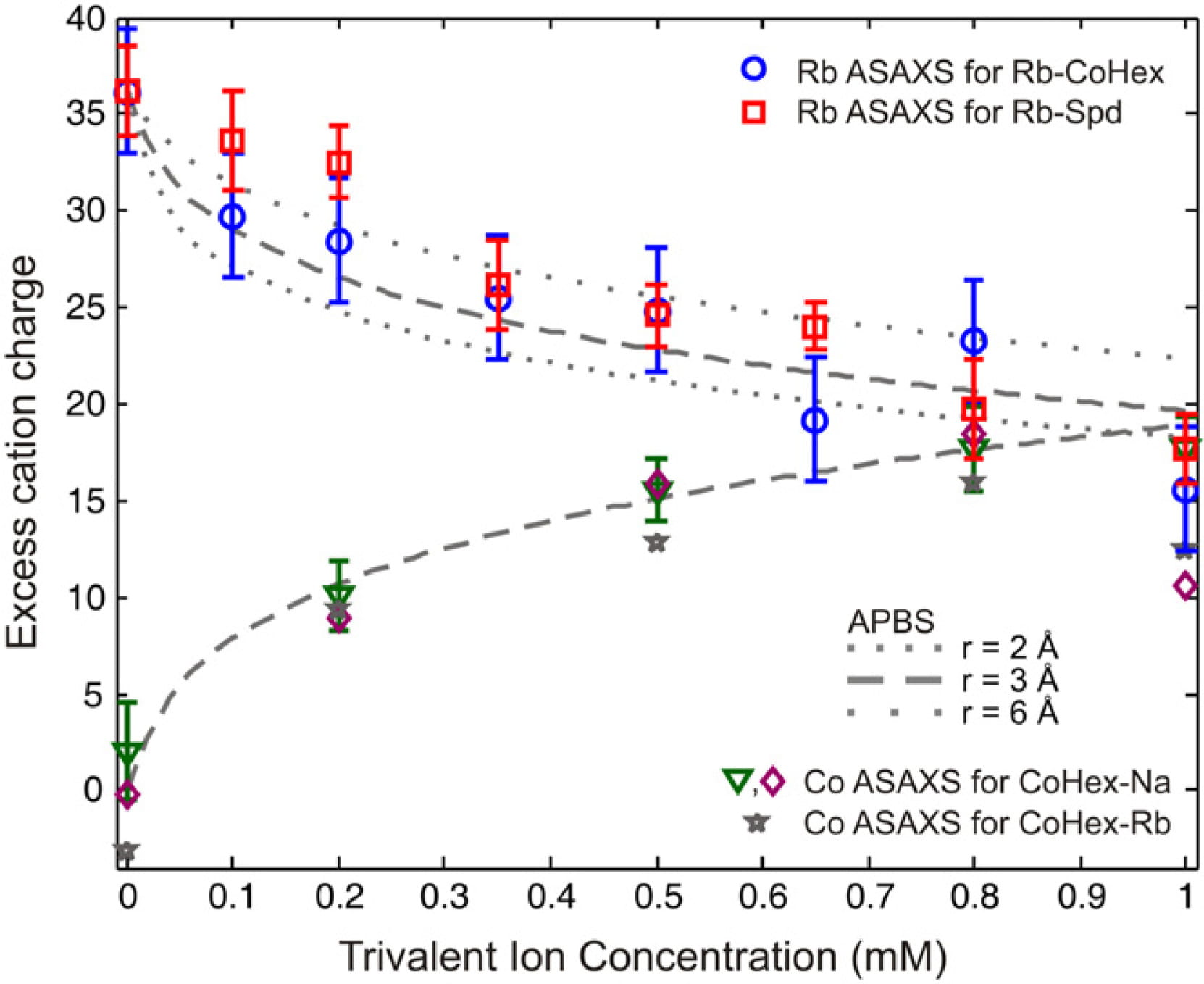
K. Andresen, X. Qiu, S. A. Pabit, J. S. Lamb, H.Y. Park, L.W. Kwok and L. Pollack
Mono and tri-valent ions around DNA: a small angle scattering study of competition and interactions.
Biophys. J., 95, 287 (2008)
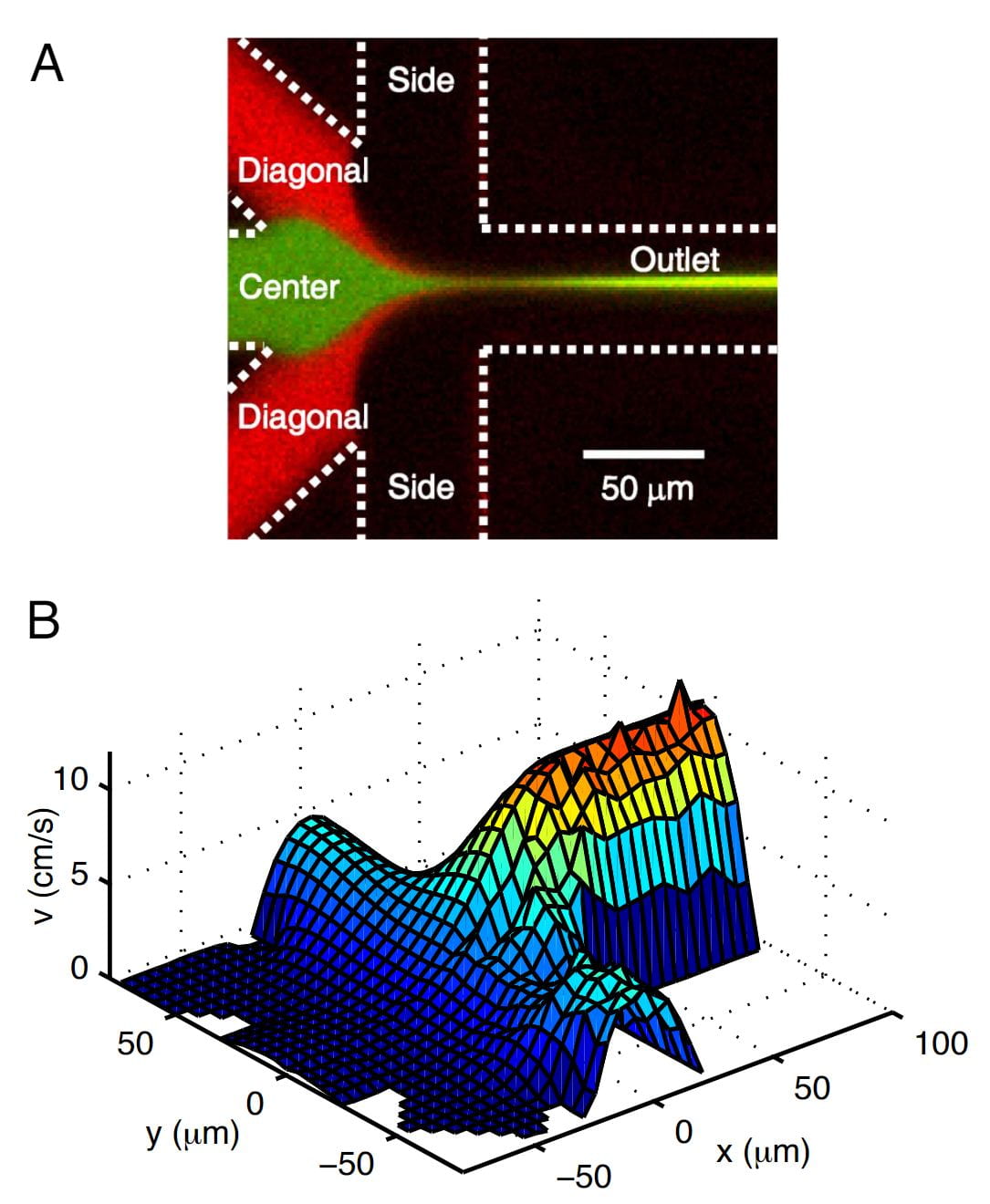
H. Y. Park, S. A. Kim, J. Korlach, E. Rhoades, L. W. Kwok, W. R. Zipfel, M. N. Waxham, W. W. Webb, and L. Pollack
Conformational changes of calmodulin upon Ca2+ binding studied with a microfluidic mixer.
Proc. Natl Acad Sci USA, 105(2), 542 (2008)
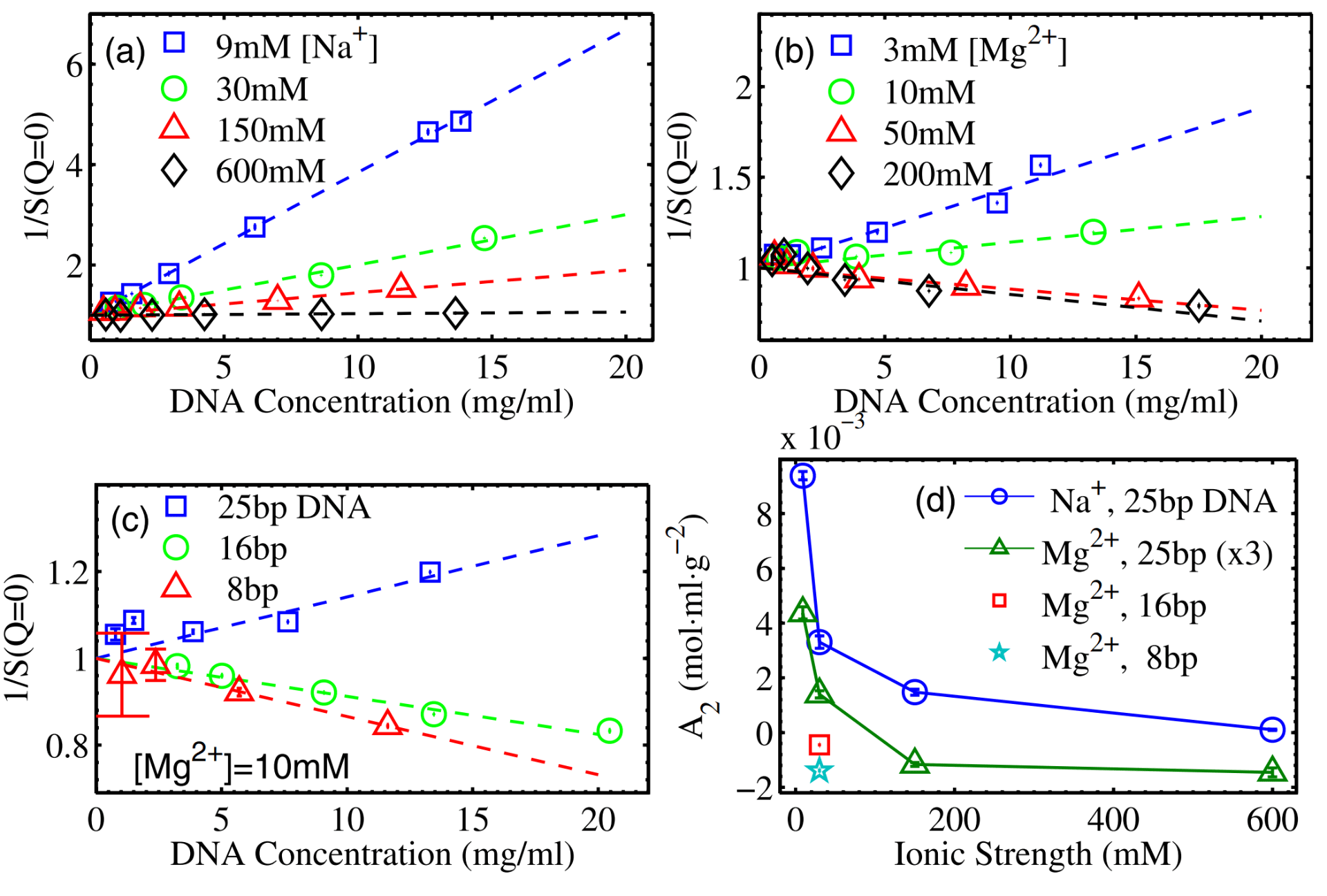
X. Qiu, K. Andresen, L.W. Kwok, J.S. Lamb, H.Y. Park, and L. Pollack
Inter-DNA attraction mediated by divalent counterions.
Phys. Rev. Lett., 99 038104 (2007)
J.S. Lamb, S. Cornaby, K. Andresen, L.W. Kwok, H.Y. Park, X. Qiu, D.M. Smilgies, D.H. Bilderback and L. Pollack,
Focusing capillary optics for use in solution SAXS.
J. Appl. Cryst., 40, 193 (2007)
H.Y. Park, X. Qiu, E. Rhoades, J. Korlach, L.W. Kwok, W.R. Zipfel, W.W. Webb and L. Pollack
Achieving uniform mixing in a microfluidic device: hydrodynamic focusing prior to mixing.
Anal. Chem., 78, 4465 (2006)
X. Qiu, L.W. Kwok, H.Y. Park, J.S. Lamb, K. Andresen, and L. Pollack
Measuring inter-DNA potentials in solution.
Phys. Rev. Lett., 96 138101 (2007)
L.W. Kwok, I. Shcherbakova, J.S. Lamb, H.Y. Park, K. Andresen, H. Smith, M. Brenowitz and L. Pollack
Concordant measurements of folding kinetics from local and global perspective.
J. Mol. Biol., 355, 282 (2006)
K. Andresen, R. Das, H.Y. Park, L.W. Kwok, J.S. Lamb, H. Smith, D. Herschlag, K.D. Finkelstein and L. Pollack
Spatial distribution of competing ions around DNA.
Phys. Rev. Lett., 93, 248103, (2004)
R. Das, T. T. Mills, L. W. Kwok, G. S. Maskel, I.S. Millett, D. Doniach, K. D. Finkelstein, D. Herschlag and L. Pollack
Counterion distribution around DNA probed by x-ray scattering.
Phys. Rev. Lett., 90, 188103 (2003)
R. Das, L.W. Kwok, I.S. Millett, Y. Bai, T.T. Mills, J. Jacob, G.S. Maskel, S. Seifert, S.G.J. Mochrie, P. Thiyagarajan, S. Doniach, L. Pollack, and D. Herschlag
The fastest global events in RNA folding.
J. Mol. Biol., 332, 311 (2003)
R. Russell, I. S. Mil.ett, M. W. Tate, L. W. Kwok, B. Nakatani, S. M. Gruner, S. G. J. Mochrine, V. Pande, S. Doniach, D. Herschlag, and L. Pollack
Rapid compaction during RNA folding.
Proc. Natl. Acad. Sci. 99, p. 4266 (2002)
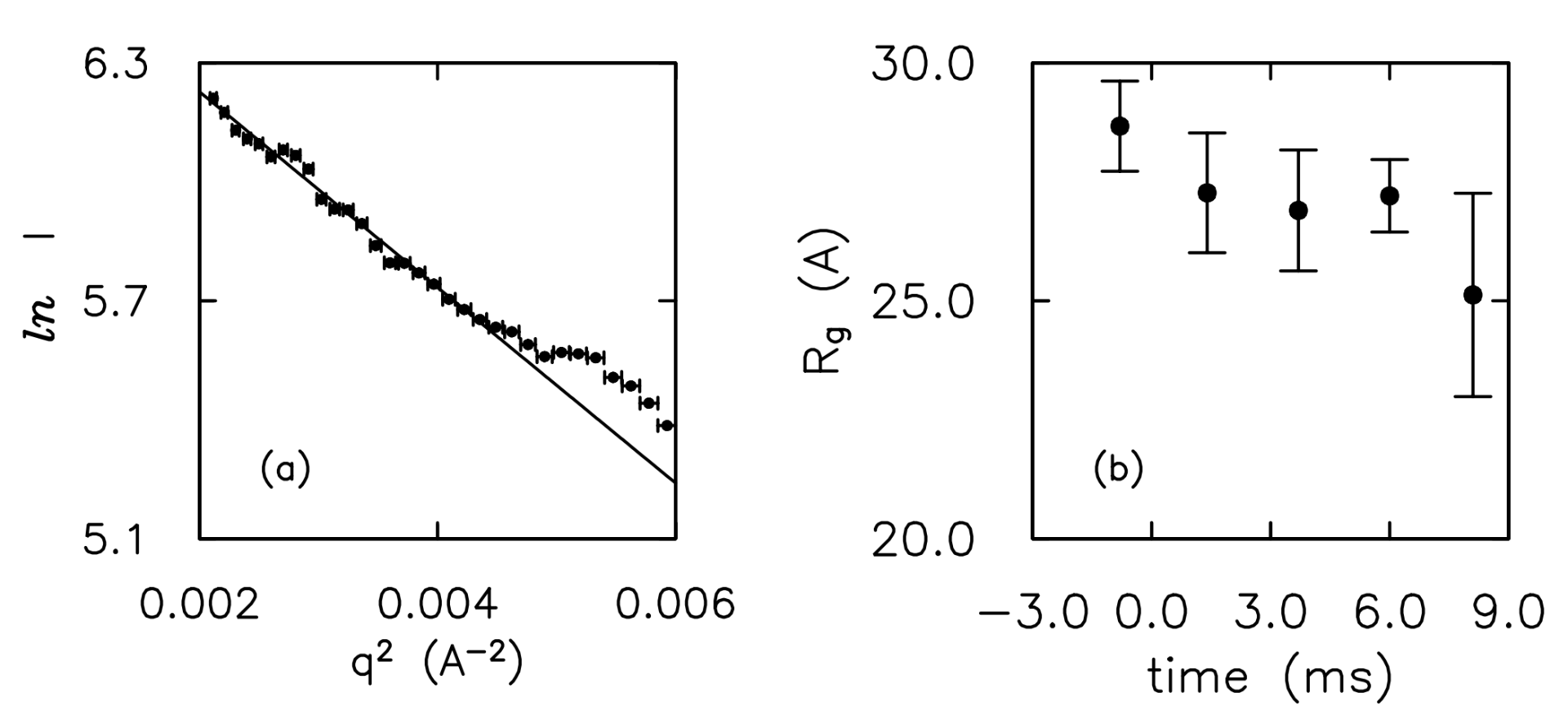
L. Pollack, M. W. Tate, A. C. Finnefrock, C. Kalidas, S. Trotter, N. C. Darnton, L. Lurio, R. H. Austin, C. A. Batt, S. M. Gruner, and S. G. J. Mochrine
Time-resolved collapse of a folding protein observed with small angle x-ray scattering.
Phys. Rev. Lett. 86, p. 4692 (2001)
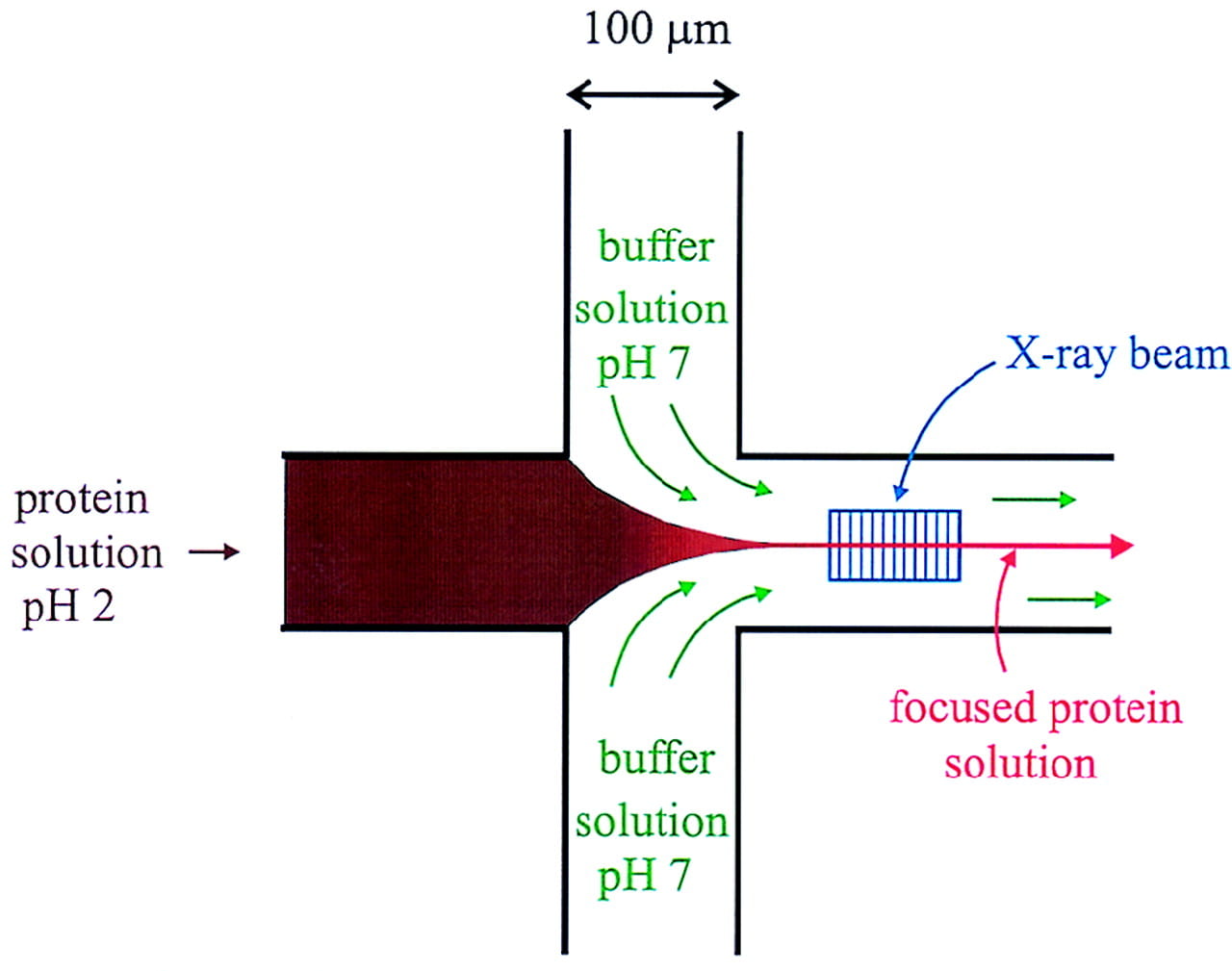
L. Pollack, M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W. A. Eaton, and R. H. Austin
Compactness of the denatured state of a fast-folding protein measured by submillisecond small angle x-ray scattering.
Proc. Natl. Acad. Sci. USA 96, 10115-10117 (1999)