Development of Microfluidic Mixers

Biology is filled with active and dynamic processes: DNA is read by protein machinery to make RNA, RNA cuts and folds itself to edit its own sequence and is later used as a template for more proteins which perform a slew of cellular functions. Capturing these processes requires a way to ‘ride along’ with the biological machinery, watching in time as they function.
Specifically, we want to see how biomolecules react to changes in solution conditions: adding magnesium ions causes RNA to collapse and fold into its most compact three dimensional structures, adding a small-molecule ligand causes structural change within an RNA riboswitch. To see how these happen in real time and probe them in the time-domain, we design, build and implement state of the art microfluidic mixing devices and dual color microscopes. This combination allows us to watch individual biological machines going about their business, providing an up close and personal look at processes as they occur.
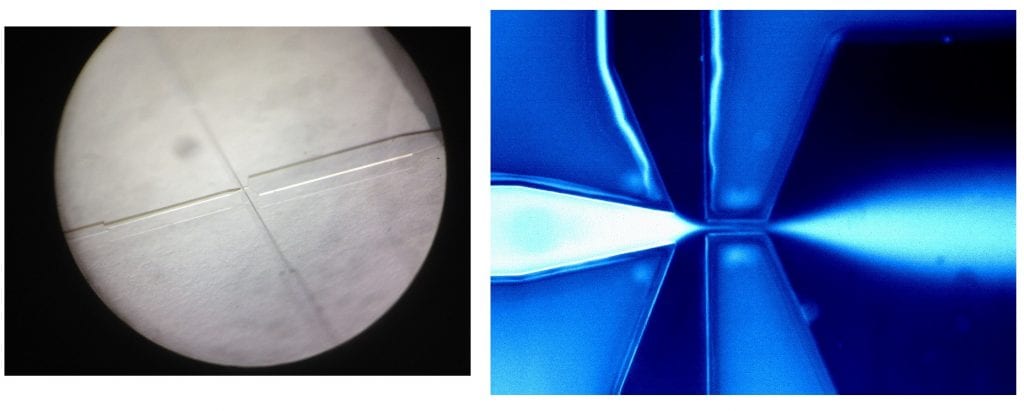
Microfluidic mixing devices are nanofabricated chips designed to rapidly mix small volumes of samples together. Mixing occurs in the chip, and the solution then delivered to a sample chamber to allow probing at varying times during the reaction (typically from milliseconds to seconds). We fabricate these devices at Cornell’s cleanroom CNF, using a variety of nanofabrication tools to form structures on the ~10μm scale. As it turns out, pajamas and big tools are sometimes required to make small things (right inset)! Our latest microfabricated chips are probed using a two-color confocal microscope system which we designed to detect single, fluorescently labelled biomolecules.
With these tools we have watched many exciting interactions from RNA gene silencing to virus assembly, a single-molecule at a time [1]!
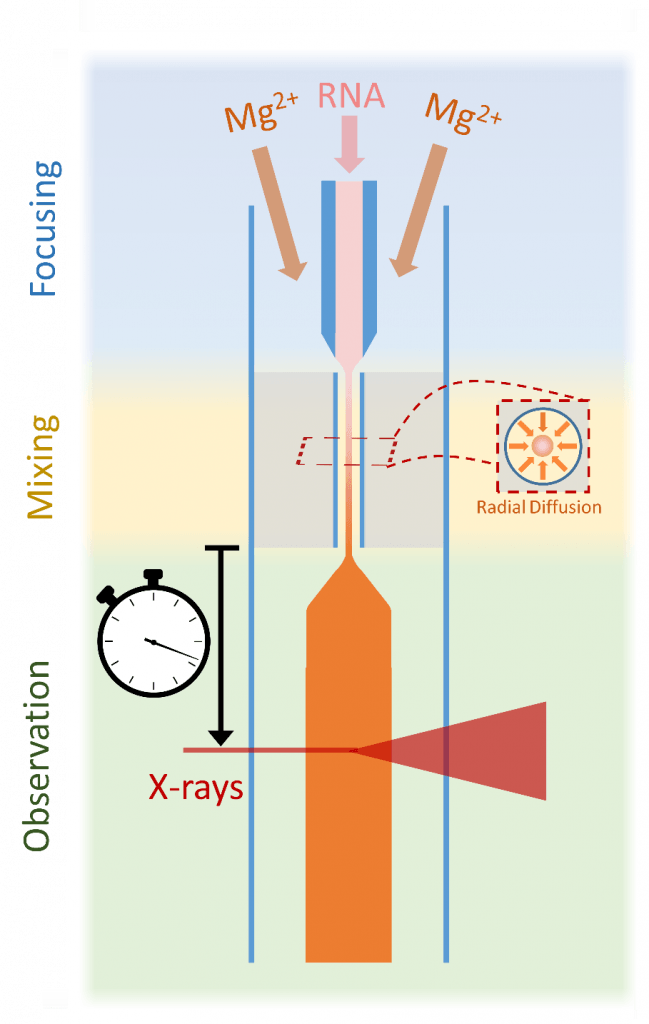
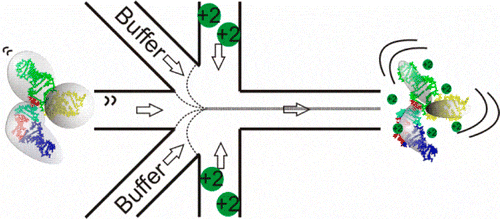
We’ve also used microfabricated microfluidic mixers to study the folding of RNA. Following the addition of ions to trigger folding, RNA molecules undergo a transition from rigid, extended states to a compact ensemble. Here, microfluidic mixing with microsecond time resolution and fluorescence resonance energy transfer (FRET) detection provides insight into the ionic strength-dependent transition from extended to compact ensembles. Differences in reaction rates are detected when folding is initiated by monovalent or divalent ions, consistent with equilibrium measurements illustrating the enhanced screening of divalent ions relative to monovalent ions at the same ionic strength. Ion-driven collapse is fast, and a comparison of the collapse times suggests that site binding of Mg2+ occurs on submillisecond time scales [2]. In the figure below, we show a schematic of the microfluidic 5-port mixer to look for studying RNA folding.
For time-resolved SAXS measurements, the Pollack Group pioneered silicon-based microfluidic devices to look into global structural changes in protein [3] and RNA molecules [4]. More recently, for SAXS-based microfluidic mixers, we have implemented capillary-based coaxial geometries to facilitate fast diffusion into the mixing region [5, 6].
References
- A. Plumridge et al. Following folding pathways of common riboswitch motifs with time-resolved single-molecule FRET. Biophysical society meeting 2019.
- S. Pabit, J. Sutton, et al. Role of Ion Valence in the Submillisecond Collapse and Folding of a Small RNA Domain, Biochemistry, 52: 1539-46 2013
- L. Pollack, et al., PNAS, 1999
- R. Russel, et al., PNAS 2002
- A. Plumridge, A. M. Katz, G. D. Calvey, et al., Nucleic Acids Research, 46, pp 7354-7365 (2018).
- R. Welty, S. Pabit, et al., RNA, 24, pp 1828-1838 (2018).