Contrast Variation Small Angle X-Ray Scattering
In the Pollack lab we use Contrast Variation (CV) Small Angle X-Ray Scattering (SAXS) to study protein nucleic acid complexes. By raising the solvent’s electron density with sucrose, this technique allows us to effectively make proteins invisible to X-Rays. In this manner we can directly observe the structure of the nucleic acids and how they interact with proteins. We have used this technique to observe DNA unwrapping from histone cores [1, 2], study protein-RNA interactions of ribosome fragments, and to look inside viruses and study their assembly.
In the lab we have developed techniques and tools to be able to use this technique in a time resolved experiment.

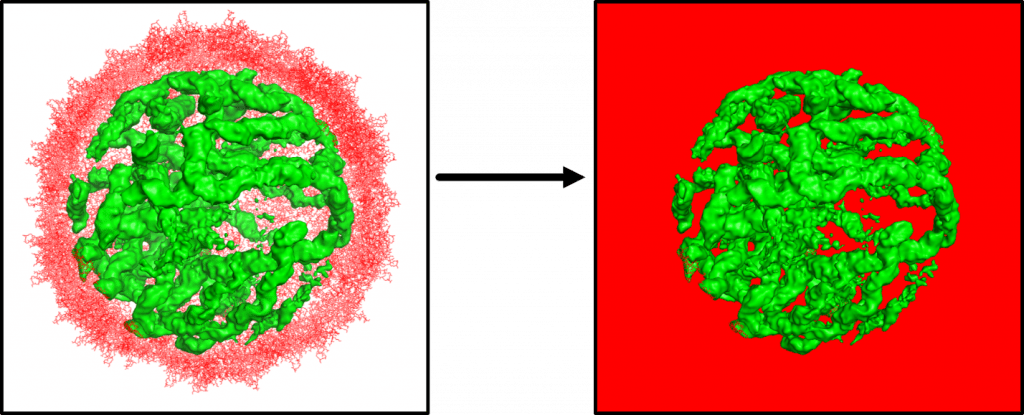
Ion-Nucleic Acid and Protein-Nucleid Acid Interactions
Nucleic acids are negatively charged and are always surrounded by a cloud of positively charged counterions. Due to careful implementation of SAXS with absolute calibration, we can determine the number and locations of these ions [2,3]. Analysis of Anomalous Small Angle X-ray Scattering measurements on DNA in conjunction with molecular modeling gives us a better understanding of nucleic acids and its ion atmosphere [4]. We are currently applying absolute calibrated SAXS measurements, together with contrast variation techniques, to fully understand interactions of RNAs and protein binding partners.

The question of RNA condensation and its link to RNA flexibility
In spite of the high negative change of nucleic acids, DNA molecules compact to small structures and gets bundled into the cell nucleus. In the Pollack Lab, we are beginning to realize that RNA molecules do not follow the same condensation mechanism that DNAs do [5]. We are using Wide-Angle X-ray Scattering techniques to figure out what happens to RNA in presence of condensing agents [6] and how it links to RNA flexibility.
References
- Y. Chen, J. Tokuda, et al., PNAS 2017
- J. Tokuda, S. Pabit, et al., Biophysical Reviews, 2016
- S. Meisburger, et al., Biophysical Journal, 2015
- H. T. Nguyen, S. A. Pabit, S. P. Meisburger, L. Pollack, D. A. Case
J. Chem. Phys. 141(22) (2014) - A. M. Katz, I. S. Tolokh, S. A. Pabit, A. V. Onufriev, L. Pollack
Biophys. J. 112(1):22-30 (2017) - S. A. Pabit, A. M. Katz, I. S. Tolokh, A. Drozdetski, N. Baker, A. V. Onufriev, L. Pollack
J. Chem. Phys. 144(20):205102 (2016)